The homeodomain complement of the ctenophore Mnemiopsis leidyi suggests that Ctenophora and Porifera diverged prior to the ParaHoxozoa
- PMID: 20920347
- PMCID: PMC2959044
- DOI: 10.1186/2041-9139-1-9
The homeodomain complement of the ctenophore Mnemiopsis leidyi suggests that Ctenophora and Porifera diverged prior to the ParaHoxozoa
Abstract
Background: The much-debated phylogenetic relationships of the five early branching metazoan lineages (Bilateria, Cnidaria, Ctenophora, Placozoa and Porifera) are of fundamental importance in piecing together events that occurred early in animal evolution. Comparisons of gene content between organismal lineages have been identified as a potentially useful methodology for phylogenetic reconstruction. However, these comparisons require complete genomes that, until now, did not exist for the ctenophore lineage. The homeobox superfamily of genes is particularly suited for these kinds of gene content comparisons, since it is large, diverse, and features a highly conserved domain.
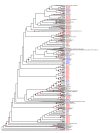
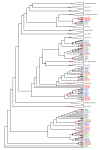
Results: We have used a next-generation sequencing approach to generate a high-quality rough draft of the genome of the ctenophore Mnemiopsis leidyi and subsequently identified a set of 76 homeobox-containing genes from this draft. We phylogenetically categorized this set into established gene families and classes and then compared this set to the homeodomain repertoire of species from the other four early branching metazoan lineages. We have identified several important classes and subclasses of homeodomains that appear to be absent from Mnemiopsis and from the poriferan Amphimedon queenslandica. We have also determined that, based on lineage-specific paralog retention and average branch lengths, it is unlikely that these missing classes and subclasses are due to extensive gene loss or unusually high rates of evolution in Mnemiopsis.
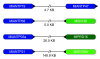
Conclusions: This paper provides a first glimpse of the first sequenced ctenophore genome. We have characterized the full complement of Mnemiopsis homeodomains from this species and have compared them to species from other early branching lineages. Our results suggest that Porifera and Ctenophora were the first two extant lineages to diverge from the rest of animals. Based on this analysis, we also propose a new name - ParaHoxozoa - for the remaining group that includes Placozoa, Cnidaria and Bilateria.
Figures


Similar articles
-
Genomic organization, evolution, and expression of photoprotein and opsin genes in Mnemiopsis leidyi: a new view of ctenophore photocytes.BMC Biol. 2012 Dec 21;10:107. doi: 10.1186/1741-7007-10-107. BMC Biol. 2012. PMID: 23259493 Free PMC article.
-
Nuclear receptors from the ctenophore Mnemiopsis leidyi lack a zinc-finger DNA-binding domain: lineage-specific loss or ancestral condition in the emergence of the nuclear receptor superfamily?Evodevo. 2011 Feb 3;2(1):3. doi: 10.1186/2041-9139-2-3. Evodevo. 2011. PMID: 21291545 Free PMC article.
-
Genomic insights into Wnt signaling in an early diverging metazoan, the ctenophore Mnemiopsis leidyi.Evodevo. 2010 Oct 4;1(1):10. doi: 10.1186/2041-9139-1-10. Evodevo. 2010. PMID: 20920349 Free PMC article.
-
Whole-Body Regeneration in the Lobate Ctenophore Mnemiopsis leidyi.Genes (Basel). 2021 Jun 5;12(6):867. doi: 10.3390/genes12060867. Genes (Basel). 2021. PMID: 34198839 Free PMC article. Review.
-
Brief History of Ctenophora.Methods Mol Biol. 2024;2757:1-26. doi: 10.1007/978-1-0716-3642-8_1. Methods Mol Biol. 2024. PMID: 38668961 Review.
Cited by
-
Coral comparative genomics reveal expanded Hox cluster in the cnidarian-bilaterian ancestor.Integr Comp Biol. 2012 Dec;52(6):835-41. doi: 10.1093/icb/ics098. Epub 2012 Jul 4. Integr Comp Biol. 2012. PMID: 22767488 Free PMC article.
-
Structural and evolutionary insights into astacin metallopeptidases.Front Mol Biosci. 2023 Jan 4;9:1080836. doi: 10.3389/fmolb.2022.1080836. eCollection 2022. Front Mol Biosci. 2023. PMID: 36685277 Free PMC article.
-
The ventral epithelium of Trichoplax adhaerens deploys in distinct patterns cells that secrete digestive enzymes, mucus or diverse neuropeptides.Biol Open. 2019 Aug 9;8(8):bio045674. doi: 10.1242/bio.045674. Biol Open. 2019. PMID: 31366453 Free PMC article.
-
The Global Invertebrate Genomics Alliance (GIGA): developing community resources to study diverse invertebrate genomes.J Hered. 2014 Jan-Feb;105(1):1-18. doi: 10.1093/jhered/est084. J Hered. 2014. PMID: 24336862 Free PMC article.
-
Calcisponges have a ParaHox gene and dynamic expression of dispersed NK homeobox genes.Nature. 2014 Oct 30;514(7524):620-3. doi: 10.1038/nature13881. Nature. 2014. PMID: 25355364
References
-
- Cuvier G. Le Règne Animal Distribué Selon son Organisation, pour Servir de Base à l'Histoire Naturelle des Animaux et d'Introduction à l'Anatomie Comparée. Paris: Deterville; 1817.
-
- Leuckart R. Ueber die Morphologie und die Verwandtschaftsverhältnisse der wirbellosen thiere. Ein Beitrag zur Charakteristik und Classification der thierischen Formen. Braunschweig,: F. Vieweg und Sohn;; 1848.
-
- Harbison GR. In: The Origins and Relationships of Lower Iinvertebrates. Conway Morris S, George JD, Gibson R, Platt HM, editor. Vol. 28. London, UK: The Systematics Assocation; 1985. On the classification and evolution of the Ctenophora; pp. 78–100.
-
- Bridge D, Cunningham CW, DeSalle R, Buss LW. Class-level relationships in the phylum Cnidaria: molecular and morphological evidence. Mol Biol Evol. 1995;12:679–689. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

