The RAD51 and DMC1 homoeologous genes of bread wheat: cloning, molecular characterization and expression analysis
- PMID: 20920212
- PMCID: PMC2962619
- DOI: 10.1186/1756-0500-3-245
The RAD51 and DMC1 homoeologous genes of bread wheat: cloning, molecular characterization and expression analysis
Abstract
Background: Meiotic recombination in eukaryotes requires two homologues of the E. coli RecA proteins: Rad51 and Dmc1. Both proteins play important roles in the binding of single stranded DNA, homology search, strand invasion and strand exchange. Meiotic recombination has been well studied in Arabidopsis, rice, maize and the orthologues of RAD51 and DMC1 have been characterized. However genetic analysis of the RAD51 and DMC1 genes in bread wheat has been hampered due to the absence of complete sequence information and because of the existence of multiple copies of each gene in the hexaploid wheat genome.
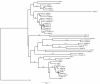
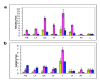
Findings: In this study we have identified that TaRAD51 and TaDMC1 homoeologues are located on group 7 and group 5 chromosomes of hexaploid wheat, respectively. Comparative sequence analysis of cDNA derived from the TaRAD51 and TaDMC1 homoeologues revealed limited sequence divergence at both the nucleotide and the amino acid level. Indeed, comparisons between the predicted amino acid sequences of TaRAD51 and TaDMC1 and those of other eukaryotes reveal a high degree of evolutionary conservation. Despite the high degree of sequence conservation at the nucleotide level, genome-specific primers for cDNAs of TaRAD51 and TaDMC1 were developed to evaluate expression patterns of individual homoeologues during meiosis. QRT-PCR analysis showed that expression of the TaRAD51 and TaDMC1 cDNA homoeologues was largely restricted to meiotic tissue, with elevated levels observed during the stages of prophase I when meiotic recombination occurs. All three homoeologues of both strand-exchange proteins (TaRAD51 and TaDMC1) are expressed in wheat.
Conclusions: Bread wheat contains three expressed copies of each of the TaRAD51 and TaDMC1 homoeologues. While differences were detected between the three cDNA homoeologues of TaRAD51 as well as the three homoeologues of TaDMC1, it is unlikely that the predicted amino acid substitutions would have an effect on the protein structure, based on our three-dimensional structure prediction analyses. There are differences in the levels of expression of the three homoeologues of TaRAD51 and TaDMC1 as determined by QRT-PCR and if these differences are reflected at the protein level, bread wheat may be more dependent upon a particular homoeologue to achieve full fertility than all three equally.
Figures





Similar articles
-
Relationship between homoeologous regulatory and structural genes in allopolyploid genome - a case study in bread wheat.BMC Plant Biol. 2008 Aug 13;8:88. doi: 10.1186/1471-2229-8-88. BMC Plant Biol. 2008. PMID: 18700978 Free PMC article.
-
The RAD51 gene family in bread wheat is highly conserved across eukaryotes, with RAD51A upregulated during early meiosis.Funct Plant Biol. 2008 Dec;35(12):1267-1277. doi: 10.1071/FP08203. Funct Plant Biol. 2008. PMID: 32688873
-
Saccharomyces cerevisiae recA homologues RAD51 and DMC1 have both distinct and overlapping roles in meiotic recombination.Genes Cells. 1997 Oct;2(10):615-29. doi: 10.1046/j.1365-2443.1997.1480347.x. Genes Cells. 1997. PMID: 9427283
-
Roles of RecA homologues Rad51 and Dmc1 during meiotic recombination.Cytogenet Genome Res. 2004;107(3-4):201-7. doi: 10.1159/000080598. Cytogenet Genome Res. 2004. PMID: 15467365 Review.
-
Repair of DNA double-strand breaks in plant meiosis: role of eukaryotic RecA recombinases and their modulators.Plant Reprod. 2023 Mar;36(1):17-41. doi: 10.1007/s00497-022-00443-6. Epub 2022 Jun 1. Plant Reprod. 2023. PMID: 35641832 Review.
Cited by
-
Morphotype transition and sexual reproduction are genetically associated in a ubiquitous environmental pathogen.PLoS Pathog. 2014 Jun 5;10(6):e1004185. doi: 10.1371/journal.ppat.1004185. eCollection 2014 Jun. PLoS Pathog. 2014. PMID: 24901238 Free PMC article.
-
On the role of AtDMC1, AtRAD51 and its paralogs during Arabidopsis meiosis.Front Plant Sci. 2014 Feb 17;5:23. doi: 10.3389/fpls.2014.00023. eCollection 2014. Front Plant Sci. 2014. PMID: 24596572 Free PMC article. Review.
-
A Co-Expression Network in Hexaploid Wheat Reveals Mostly Balanced Expression and Lack of Significant Gene Loss of Homeologous Meiotic Genes Upon Polyploidization.Front Plant Sci. 2019 Oct 18;10:1325. doi: 10.3389/fpls.2019.01325. eCollection 2019. Front Plant Sci. 2019. PMID: 31681395 Free PMC article.
-
Homologous pairing activities of two rice RAD51 proteins, RAD51A1 and RAD51A2.PLoS One. 2013 Oct 4;8(10):e75451. doi: 10.1371/journal.pone.0075451. eCollection 2013. PLoS One. 2013. PMID: 24124491 Free PMC article.
-
Dmc1 is a candidate for temperature tolerance during wheat meiosis.Theor Appl Genet. 2020 Mar;133(3):809-828. doi: 10.1007/s00122-019-03508-9. Epub 2019 Dec 18. Theor Appl Genet. 2020. PMID: 31853574 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

