DAnCER: disease-annotated chromatin epigenetics resource
- PMID: 20876685
- PMCID: PMC3013761
- DOI: 10.1093/nar/gkq857
DAnCER: disease-annotated chromatin epigenetics resource
Abstract
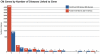
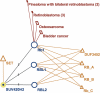
Chromatin modification (CM) is a set of epigenetic processes that govern many aspects of DNA replication, transcription and repair. CM is carried out by groups of physically interacting proteins, and their disruption has been linked to a number of complex human diseases. CM remains largely unexplored, however, especially in higher eukaryotes such as human. Here we present the DAnCER resource, which integrates information on genes with CM function from five model organisms, including human. Currently integrated are gene functional annotations, Pfam domain architecture, protein interaction networks and associated human diseases. Additional supporting evidence includes orthology relationships across organisms, membership in protein complexes, and information on protein 3D structure. These data are available for 962 experimentally confirmed and manually curated CM genes and for over 5000 genes with predicted CM function on the basis of orthology and domain composition. DAnCER allows visual explorations of the integrated data and flexible query capabilities using a variety of data filters. In particular, disease information and functional annotations are mapped onto the protein interaction networks, enabling the user to formulate new hypotheses on the function and disease associations of a given gene based on those of its interaction partners. DAnCER is freely available at http://wodaklab.org/dancer/.
Figures
Similar articles
-
BioGRID: a general repository for interaction datasets.Nucleic Acids Res. 2006 Jan 1;34(Database issue):D535-9. doi: 10.1093/nar/gkj109. Nucleic Acids Res. 2006. PMID: 16381927 Free PMC article.
-
Genome-wide prediction of C. elegans genetic interactions.Science. 2006 Mar 10;311(5766):1481-4. doi: 10.1126/science.1123287. Science. 2006. PMID: 16527984
-
The BioGRID Interaction Database: 2008 update.Nucleic Acids Res. 2008 Jan;36(Database issue):D637-40. doi: 10.1093/nar/gkm1001. Epub 2007 Nov 13. Nucleic Acids Res. 2008. PMID: 18000002 Free PMC article.
-
Lessons from modENCODE.Annu Rev Genomics Hum Genet. 2015;16:31-53. doi: 10.1146/annurev-genom-090413-025448. Epub 2015 Jun 26. Annu Rev Genomics Hum Genet. 2015. PMID: 26133010 Review.
-
Genetics of eukaryotic RNA polymerases I, II, and III.Microbiol Rev. 1993 Sep;57(3):703-24. doi: 10.1128/mr.57.3.703-724.1993. Microbiol Rev. 1993. PMID: 8246845 Free PMC article. Review.
Cited by
-
Network Approaches for Charting the Transcriptomic and Epigenetic Landscape of the Developmental Origins of Health and Disease.Genes (Basel). 2022 Apr 26;13(5):764. doi: 10.3390/genes13050764. Genes (Basel). 2022. PMID: 35627149 Free PMC article. Review.
-
MIAOME: Human microbiome affect the host epigenome.Comput Struct Biotechnol J. 2022 May 17;20:2455-2463. doi: 10.1016/j.csbj.2022.05.024. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35664224 Free PMC article.
-
Genetic, Epigenetic, and Steroidogenic Modulation Mechanisms in Endometriosis.J Clin Med. 2020 May 2;9(5):1309. doi: 10.3390/jcm9051309. J Clin Med. 2020. PMID: 32370117 Free PMC article. Review.
-
HEMD: an integrated tool of human epigenetic enzymes and chemical modulators for therapeutics.PLoS One. 2012;7(6):e39917. doi: 10.1371/journal.pone.0039917. Epub 2012 Jun 25. PLoS One. 2012. PMID: 22761927 Free PMC article.
-
Savant Genome Browser 2: visualization and analysis for population-scale genomics.Nucleic Acids Res. 2012 Jul;40(Web Server issue):W615-21. doi: 10.1093/nar/gks427. Epub 2012 May 25. Nucleic Acids Res. 2012. PMID: 22638571 Free PMC article.
References
-
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. - PubMed
-
- Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128:721–733. - PubMed
-
- Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat. Rev. Cancer. 2006;6:38–51. - PubMed
-
- Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. - PubMed
-
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

