A QUICK screen for Lrrk2 interaction partners--leucine-rich repeat kinase 2 is involved in actin cytoskeleton dynamics
- PMID: 20876399
- PMCID: PMC3013447
- DOI: 10.1074/mcp.M110.001172
A QUICK screen for Lrrk2 interaction partners--leucine-rich repeat kinase 2 is involved in actin cytoskeleton dynamics
Abstract
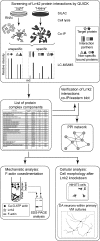
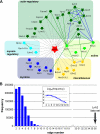
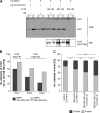
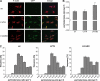
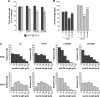
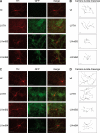
Mutations in human leucine-rich repeat kinase 2 (Lrrk2), a protein of yet unknown function, are linked to Parkinson's disease caused by degeneration of midbrain dopaminergic neurons. The protein comprises several domains including a GTPase and a kinase domain both affected by several pathogenic mutations. To elucidate the molecular interaction network of endogenous Lrrk2 under stoichiometric constraints, we applied QUICK (quantitative immunoprecipitation combined with knockdown) in NIH3T3 cells. The identified interactome reveals actin isoforms as well as actin-associated proteins involved in actin filament assembly, organization, rearrangement, and maintenance, suggesting that the biological function of Lrrk2 is linked to cytoskeletal dynamics. In fact, we demonstrate Lrrk2 de novo binding to F-actin and its ability to modulate its assembly in vitro. When tested in intact cells, knockdown of Lrrk2 causes morphological alterations in NIH3T3 cells. In developing dopaminergic midbrain primary neurons, Lrrk2 knockdown results in shortened neurite processes, indicating a physiological role of Lrrk2 in cytoskeletal organization and dynamics of dopaminergic neurons. Hence, our results demonstrate that molecular interactions as well as the physiological function of Lrrk2 are closely related to the organization of the actin-based cytoskeleton, a crucial feature of neuronal development and neuron function.
Figures

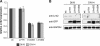
Similar articles
-
LRRK2 guides the actin cytoskeleton at growth cones together with ARHGEF7 and Tropomyosin 4.Biochim Biophys Acta. 2013 Dec;1832(12):2352-67. doi: 10.1016/j.bbadis.2013.09.009. Epub 2013 Sep 24. Biochim Biophys Acta. 2013. PMID: 24075941
-
Phosphorylation of ezrin/radixin/moesin proteins by LRRK2 promotes the rearrangement of actin cytoskeleton in neuronal morphogenesis.J Neurosci. 2009 Nov 4;29(44):13971-80. doi: 10.1523/JNEUROSCI.3799-09.2009. J Neurosci. 2009. PMID: 19890007 Free PMC article.
-
Rac1 protein rescues neurite retraction caused by G2019S leucine-rich repeat kinase 2 (LRRK2).J Biol Chem. 2011 May 6;286(18):16140-9. doi: 10.1074/jbc.M111.234005. Epub 2011 Mar 16. J Biol Chem. 2011. PMID: 21454543 Free PMC article.
-
On the road to leucine-rich repeat kinase 2 signalling: evidence from cellular and in vivo studies.Neurosignals. 2011;19(1):1-15. doi: 10.1159/000324488. Epub 2011 Mar 23. Neurosignals. 2011. PMID: 21430363 Review.
-
[Molecular basis of Parkinson's disease linked with mutations in the LRRK2 gene].Mol Biol (Mosk). 2014 Jan-Feb;48(1):3-14. Mol Biol (Mosk). 2014. PMID: 25842821 Review. Russian.
Cited by
-
LRRK2 kinase inhibition prevents pathological microglial phagocytosis in response to HIV-1 Tat protein.J Neuroinflammation. 2012 Nov 29;9:261. doi: 10.1186/1742-2094-9-261. J Neuroinflammation. 2012. PMID: 23190742 Free PMC article.
-
GTPase activity and neuronal toxicity of Parkinson's disease-associated LRRK2 is regulated by ArfGAP1.PLoS Genet. 2012;8(2):e1002526. doi: 10.1371/journal.pgen.1002526. Epub 2012 Feb 9. PLoS Genet. 2012. PMID: 22363216 Free PMC article.
-
Cellular processes associated with LRRK2 function and dysfunction.FEBS J. 2015 Aug;282(15):2806-26. doi: 10.1111/febs.13305. Epub 2015 May 9. FEBS J. 2015. PMID: 25899482 Free PMC article. Review.
-
Roco Proteins and the Parkinson's Disease-Associated LRRK2.Int J Mol Sci. 2018 Dec 17;19(12):4074. doi: 10.3390/ijms19124074. Int J Mol Sci. 2018. PMID: 30562929 Free PMC article. Review.
-
A proteomic analysis of differentiating dopamine neurons derived from human embryonic stem cells.Anim Cells Syst (Seoul). 2019 Apr 11;23(3):219-227. doi: 10.1080/19768354.2019.1595140. eCollection 2019 Jun. Anim Cells Syst (Seoul). 2019. PMID: 31231586 Free PMC article.
References
-
- Elbaz A. (2008) LRRK2: bridging the gap between sporadic and hereditary Parkinson's disease. Lancet Neurol. 7, 562–564 - PubMed
-
- Paisán-Ruiz C. (2009) LRRK2 gene variation and its contribution to Parkinson disease. Hum. Mutat. 30, 1153–1160 - PubMed
-
- Wszolek Z. K., Pfeiffer R. F., Tsuboi Y., Uitti R. J., McComb R. D., Stoessl A. J., Strongosky A. J., Zimprich A., Müller-Myhsok B., Farrer M. J., Gasser T., Calne D. B., Dickson D. W. (2004) Autosomal dominant parkinsonism associated with variable synuclein and tau pathology. Neurology 62, 1619–1622 - PubMed
-
- Zimprich A., Biskup S., Leitner P., Lichtner P., Farrer M., Lincoln S., Kachergus J., Hulihan M., Uitti R. J., Calne D. B., Stoessl A. J., Pfeiffer R. F., Patenge N., Carbajal I. C., Vieregge P., Asmus F., Müller-Myhsok B., Dickson D. W., Meitinger T., Strom T. M., Wszolek Z. K., Gasser T. (2004) Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron 44, 601–607 - PubMed
-
- Taylor J. P., Mata I. F., Farrer M. J. (2006) LRRK2: a common pathway for parkinsonism, pathogenesis and prevention? Trends Mol. Med. 12, 76–82 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

