Identification and functional analysis of NOL7 nuclear and nucleolar localization signals
- PMID: 20875127
- PMCID: PMC2957388
- DOI: 10.1186/1471-2121-11-74
Identification and functional analysis of NOL7 nuclear and nucleolar localization signals
Abstract
Background: NOL7 is a candidate tumor suppressor that localizes to a chromosomal region 6p23. This locus is frequently lost in a number of malignancies, and consistent loss of NOL7 through loss of heterozygosity and decreased mRNA and protein expression has been observed in tumors and cell lines. Reintroduction of NOL7 into cells resulted in significant suppression of in vivo tumor growth and modulation of the angiogenic phenotype. Further, NOL7 was observed to localize to the nucleus and nucleolus of cells. However, the mechanisms regulating its subcellular localization have not been elucidated.
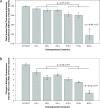
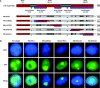
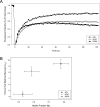
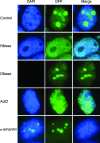
Results: An in vitro import assay demonstrated that NOL7 requires cytosolic machinery for active nuclear transport. Using sequence homology and prediction algorithms, four putative nuclear localization signals (NLSs) were identified. NOL7 deletion constructs and cytoplasmic pyruvate kinase (PK) fusion proteins confirmed the functionality of three of these NLSs. Site-directed mutagenesis of PK fusions and full-length NOL7 defined the minimal functional regions within each NLS. Further characterization revealed that NLS2 and NLS3 were critical for both the rate and efficiency of nuclear targeting. In addition, four basic clusters within NLS2 and NLS3 were independently capable of nucleolar targeting. The nucleolar occupancy of NOL7 revealed a complex balance of rapid nucleoplasmic shuttling but low nucleolar mobility, suggesting NOL7 may play functional roles in both compartments. In support, targeting to the nucleolar compartment was dependent on the presence of RNA, as depletion of total RNA or rRNA resulted in a nucleoplasmic shift of NOL7.
Conclusions: These results identify the minimal sequences required for the active targeting of NOL7 to the nucleus and nucleolus. Further, this work characterizes the relative contribution of each sequence to NOL7 nuclear and nucleolar dynamics, the subnuclear constituents that participate in this targeting, and suggests a functional role for NOL7 in both compartments. Taken together, these results identify the requisite protein domains for NOL7 localization, the kinetics that drive this targeting, and suggest NOL7 may function in both the nucleus and nucleolus.
Figures


Similar articles
-
The dynamics of the alternatively spliced NOL7 gene products and role in nucleolar architecture.Nucleus. 2011 May-Jun;2(3):229-45. doi: 10.4161/nucl.2.3.15893. Nucleus. 2011. PMID: 21818416 Free PMC article.
-
Identification and characterization of nuclear and nucleolar localization signals in 58-kDa microspherule protein (MSP58).J Biomed Sci. 2015 May 16;22(1):33. doi: 10.1186/s12929-015-0136-0. J Biomed Sci. 2015. PMID: 25981436 Free PMC article.
-
Nuclear and nucleolar localization signals and their targeting function in phosphatidylinositol 4-kinase PI4K230.Exp Cell Res. 2008 Aug 1;314(13):2376-88. doi: 10.1016/j.yexcr.2008.05.006. Epub 2008 May 27. Exp Cell Res. 2008. PMID: 18585705
-
Nucleolar-cytoplasmic shuttling of PRRSV nucleocapsid protein: a simple case of molecular mimicry or the complex regulation by nuclear import, nucleolar localization and nuclear export signal sequences.Virus Res. 2003 Sep;95(1-2):23-33. doi: 10.1016/s0168-1702(03)00161-8. Virus Res. 2003. PMID: 12921993 Free PMC article. Review.
-
Nuclear dynamics: Formation of bodies and trafficking in plant nuclei.Front Plant Sci. 2022 Aug 23;13:984163. doi: 10.3389/fpls.2022.984163. eCollection 2022. Front Plant Sci. 2022. PMID: 36082296 Free PMC article. Review.
Cited by
-
The dynamics of the alternatively spliced NOL7 gene products and role in nucleolar architecture.Nucleus. 2011 May-Jun;2(3):229-45. doi: 10.4161/nucl.2.3.15893. Nucleus. 2011. PMID: 21818416 Free PMC article.
-
Human nucleolar protein 7 (NOL7) is required for early pre-rRNA accumulation and pre-18S rRNA processing.RNA Biol. 2023 Jan;20(1):257-271. doi: 10.1080/15476286.2023.2217392. RNA Biol. 2023. PMID: 37246770 Free PMC article.
-
Pancancer Analysis of the Oncogenic and Prognostic Role of NOL7: A Potential Target for Carcinogenesis and Survival.Int J Mol Sci. 2022 Aug 25;23(17):9611. doi: 10.3390/ijms23179611. Int J Mol Sci. 2022. PMID: 36077008 Free PMC article.
-
PQBP3 prevents senescence by suppressing PSME3-mediated proteasomal Lamin B1 degradation.EMBO J. 2024 Sep;43(18):3968-3999. doi: 10.1038/s44318-024-00192-4. Epub 2024 Aug 5. EMBO J. 2024. PMID: 39103492 Free PMC article.
-
Gain of Chromosome 6p Correlates with Severe Anaplasia, Cellular Hyperchromasia, and Extraocular Spread of Retinoblastoma.Ophthalmol Sci. 2021 Dec 11;2(1):100089. doi: 10.1016/j.xops.2021.100089. eCollection 2022 Mar. Ophthalmol Sci. 2021. PMID: 36246172 Free PMC article.
References
-
- Lung ML, Choi CV, Kong H, Yuen PW, Kwong D, Sham J, Wei WI. Microsatellite allelotyping of chinese nasopharyngeal carcinomas. Anticancer Res. 2001;21(4B):3081–3084. - PubMed
-
- Lim G, Karaskova J, Vukovic B, Bayani J, Beheshti B, Bernardini M, Squire JA, Zielenska M. Combined spectral karyotyping, multicolor banding, and microarray comparative genomic hybridization analysis provides a detailed characterization of complex structural chromosomal rearrangements associated with gene amplification in the osteosarcoma cell line MG-63. Cancer Genet Cytogenet. 2004;153(2):158–164. doi: 10.1016/j.cancergencyto.2004.01.016. - DOI - PubMed
-
- Takeshita A, Naito K, Shinjo K, Sahara N, Matsui H, Ohnishi K, Beppu H, Ohtsubo K, Horii T, Maekawa M. et al.Deletion 6p23 and add(11)(p15) leading to NUP98 translocation in a case of therapy-related atypical chronic myelocytic leukemia transforming to acute myelocytic leukemia. Cancer Genet Cytogenet. 2004;152(1):56–60. doi: 10.1016/j.cancergencyto.2003.10.002. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

