Inhibition of chromatin remodeling by polycomb group protein posterior sex combs is mechanistically distinct from nucleosome binding
- PMID: 20873869
- PMCID: PMC3037448
- DOI: 10.1021/bi100532a
Inhibition of chromatin remodeling by polycomb group protein posterior sex combs is mechanistically distinct from nucleosome binding
Abstract
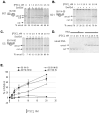
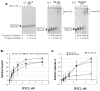
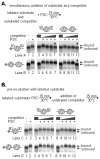
Polycomb Group (PcG) proteins are essential regulators of development that maintain gene silencing in Drosophila and mammals through alterations of chromatin structure. One key PcG protein, Posterior Sex Combs (PSC), is part of at least two complexes: Polycomb Repressive Complex 1 (PRC1) and dRING-Associated Factors (dRAF). PRC1-class complexes compact chromatin and inhibit chromatin remodeling, while dRAF has E3 ligase activity for ubiquitylation of histone H2A; activities of both complexes can inhibit transcription. The noncovalent effects of PRC1-class complexes on chromatin can be recapitulated by PSC alone, and the region of PSC required for these activities is essential for PSC function in vivo. To understand how PSC interacts with chromatin to exert its repressive effects, we compared the ability of PSC to bind to and inhibit remodeling of various nucleosomal templates and determined which regions of PSC are required for mononucleosome binding and inhibition of chromatin remodeling. We find that PSC binds mononucleosome templates but inhibits their remodeling poorly. Addition of linker DNA to mononucleosomes allows their remodeling to be inhibited, although higher concentrations of PSC are required than for inhibition of multinucleosome templates. The C-terminal region of PSC (amino acids 456−1603) is important for inhibition of chromatin remodeling, and we identified amino acids 456−909 as being sufficient for stable nucleosome binding but not for inhibition of chromatin remodeling. Our data suggest distinct mechanistic steps between nucleosome binding and inhibition of chromatin remodeling.
Figures

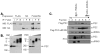
Similar articles
-
DNA Binding Reorganizes the Intrinsically Disordered C-Terminal Region of PSC in Drosophila PRC1.J Mol Biol. 2020 Aug 7;432(17):4856-4871. doi: 10.1016/j.jmb.2020.07.002. Epub 2020 Jul 3. J Mol Biol. 2020. PMID: 32628956 Free PMC article.
-
The role of the histone H2A ubiquitinase Sce in Polycomb repression.Development. 2012 Jan;139(1):117-27. doi: 10.1242/dev.074450. Epub 2011 Nov 17. Development. 2012. PMID: 22096074 Free PMC article.
-
Chromatin modification by PSC occurs at one PSC per nucleosome and does not require the acidic patch of histone H2A.PLoS One. 2012;7(10):e47162. doi: 10.1371/journal.pone.0047162. Epub 2012 Oct 11. PLoS One. 2012. PMID: 23071745 Free PMC article.
-
Chromatin regulation: how complex does it get?Epigenetics. 2014 Nov;9(11):1485-95. doi: 10.4161/15592294.2014.971580. Epigenetics. 2014. PMID: 25482055 Free PMC article. Review.
-
Polycomb response elements and targeting of Polycomb group proteins in Drosophila.Curr Opin Genet Dev. 2006 Oct;16(5):476-84. doi: 10.1016/j.gde.2006.08.005. Epub 2006 Aug 17. Curr Opin Genet Dev. 2006. PMID: 16914306 Review.
Cited by
-
DNA Binding Reorganizes the Intrinsically Disordered C-Terminal Region of PSC in Drosophila PRC1.J Mol Biol. 2020 Aug 7;432(17):4856-4871. doi: 10.1016/j.jmb.2020.07.002. Epub 2020 Jul 3. J Mol Biol. 2020. PMID: 32628956 Free PMC article.
-
Polycomb Group (PcG) Proteins and Human Cancers: Multifaceted Functions and Therapeutic Implications.Med Res Rev. 2015 Nov;35(6):1220-67. doi: 10.1002/med.21358. Epub 2015 Jul 30. Med Res Rev. 2015. PMID: 26227500 Free PMC article. Review.
-
Roles of Polycomb complexes in regulating gene expression and chromatin structure in plants.Plant Commun. 2021 Nov 26;3(1):100267. doi: 10.1016/j.xplc.2021.100267. eCollection 2022 Jan 10. Plant Commun. 2021. PMID: 35059633 Free PMC article. Review.
-
An Unexpected Regulatory Cascade Governs a Core Function of the Drosophila PRC1 Chromatin Protein Su(z)2.Genetics. 2017 Feb;205(2):551-558. doi: 10.1534/genetics.116.187849. Epub 2016 Nov 23. Genetics. 2017. PMID: 27881472 Free PMC article.
-
Ubiquitin-Specific Proteases: Players in Cancer Cellular Processes.Pharmaceuticals (Basel). 2021 Aug 26;14(9):848. doi: 10.3390/ph14090848. Pharmaceuticals (Basel). 2021. PMID: 34577547 Free PMC article. Review.
References
-
- Jurgens G. A group of genes controlling the spatial expression of the bithorax complex in Drosophila. Nature. 1985;316:153–155.
-
- Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276(5688):565–570. - PubMed
-
- Schuettengruber B, et al. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128(4):735–745. - PubMed
-
- Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. - PubMed
-
- Heard E, Disteche CM. Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev. 2006;20(14):1848–1867. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

