Association of Tat with promoters of PTEN and PP2A subunits is key to transcriptional activation of apoptotic pathways in HIV-infected CD4+ T cells
- PMID: 20862322
- PMCID: PMC2940756
- DOI: 10.1371/journal.ppat.1001103
Association of Tat with promoters of PTEN and PP2A subunits is key to transcriptional activation of apoptotic pathways in HIV-infected CD4+ T cells
Abstract
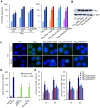
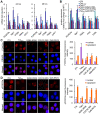
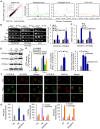
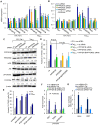
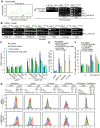
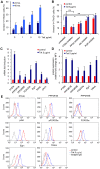
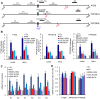
Apoptosis in HIV-1-infected CD4+ primary T cells is triggered by the alteration of the PI3K and p53 pathways, which converge on the FOXO3a transcriptional activator. Tat alone can cause activation of FOXO3a and of its proapoptotic target genes. To understand how Tat affects this pathway, we carried out ChIP-Chip experiments with Tat. Tat associates with the promoters of PTEN and two PP2A subunit genes, but not with the FOXO3a promoter. PTEN and PP2A encode phosphatases, whose levels and activity are increased when Tat is expressed. They counteract phosphorylation of Akt1 and FOXO3a, and so activate transcriptional activity of FOXO3a. FOXO3a promotes increased transcription of Egr-1, which can further stimulate the transcription of PTEN, thereby reinforcing the pathway that leads to FOXO3a transcriptional activation. RNAi experiments support the role of PTEN and PP2A in the initiation of the Tat-mediated cascade, which is critical to apoptosis. The increased accumulation of PTEN and PP2A subunit mRNAs during Tat expression is more likely to be the result of increased transcription initiation and not relief of promoter-proximal pausing of RNAPII. The Tat-PTEN and -PP2A promoter interactions provide a mechanistic explanation of Tat-mediated apoptosis in CD4+ T cells.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Foxo3a: an integrator of immune dysfunction during HIV infection.Cytokine Growth Factor Rev. 2012 Aug-Oct;23(4-5):215-21. doi: 10.1016/j.cytogfr.2012.05.008. Epub 2012 Jun 27. Cytokine Growth Factor Rev. 2012. PMID: 22748238 Free PMC article. Review.
-
Tat-induced FOXO3a is a key mediator of apoptosis in HIV-1-infected human CD4+ T lymphocytes.J Immunol. 2008 Dec 15;181(12):8460-77. doi: 10.4049/jimmunol.181.12.8460. J Immunol. 2008. PMID: 19050264 Free PMC article.
-
beta1-Integrin-collagen interaction suppresses FoxO3a by the coordination of Akt and PP2A.J Biol Chem. 2010 May 7;285(19):14195-209. doi: 10.1074/jbc.M109.052845. Epub 2010 Mar 11. J Biol Chem. 2010. PMID: 20223831 Free PMC article.
-
Involvement of the PTEN-AKT-FOXO3a pathway in neuronal apoptosis in developing rat brain after hypoxia-ischemia.J Cereb Blood Flow Metab. 2009 Dec;29(12):1903-13. doi: 10.1038/jcbfm.2009.102. Epub 2009 Jul 22. J Cereb Blood Flow Metab. 2009. PMID: 19623194 Free PMC article.
-
Modulation of apoptosis and viral latency - an axis to be well understood for successful cure of human immunodeficiency virus.J Gen Virol. 2016 Apr;97(4):813-824. doi: 10.1099/jgv.0.000402. Epub 2016 Jan 13. J Gen Virol. 2016. PMID: 26764023 Review.
Cited by
-
Foxo3a: an integrator of immune dysfunction during HIV infection.Cytokine Growth Factor Rev. 2012 Aug-Oct;23(4-5):215-21. doi: 10.1016/j.cytogfr.2012.05.008. Epub 2012 Jun 27. Cytokine Growth Factor Rev. 2012. PMID: 22748238 Free PMC article. Review.
-
Deployment of the human immunodeficiency virus type 1 protein arsenal: combating the host to enhance viral transcription and providing targets for therapeutic development.J Gen Virol. 2012 Jun;93(Pt 6):1151-1172. doi: 10.1099/vir.0.041186-0. Epub 2012 Mar 14. J Gen Virol. 2012. PMID: 22422068 Free PMC article.
-
The HIV-1 Tat protein recruits a ubiquitin ligase to reorganize the 7SK snRNP for transcriptional activation.Elife. 2018 May 30;7:e31879. doi: 10.7554/eLife.31879. Elife. 2018. PMID: 29845934 Free PMC article.
-
HIV-1 Transactivator of Transcription (Tat) Co-operates With AP-1 Factors to Enhance c-MYC Transcription.Front Cell Dev Biol. 2021 Jun 30;9:693706. doi: 10.3389/fcell.2021.693706. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34277639 Free PMC article.
-
T-Cell Signaling in HIV-1 Infection.Open Virol J. 2013 Jul 26;7:57-71. doi: 10.2174/1874357920130621001. eCollection 2013. Open Virol J. 2013. PMID: 23986795 Free PMC article.
References
-
- Modur V, Nagarajan R, Evers Bm, Milbrandt J. Foxo Proteins Regulate Tumor Necrosis Factor-Related Apoptosis Inducing Ligand Expression. Implications For Pten Mutation In Prostate Cancer. J Biol Chem. 2002;277:47928–47937. - PubMed
-
- Sunters A, Fernandez De Mattos S, Stahl M, Brosens Jj, Zoumpoulidou G, et al. Foxo3a Transcriptional Regulation Of Bim Controls Apoptosis In Paclitaxel-Treated Breast Cancer Cell Lines. J Biol Chem. 2003;278:49795–49805. - PubMed
-
- Virolle T, Adamson Ed, Baron V, Birle D, Mercola D, et al. The Egr-1 Transcription Factor Directly Activates Pten During Irradiation-Induced Signalling. Nat Cell Biol. 2001;3:1124–1128. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

