The ploidy conveyor of mature hepatocytes as a source of genetic variation
- PMID: 20861837
- PMCID: PMC2967727
- DOI: 10.1038/nature09414
The ploidy conveyor of mature hepatocytes as a source of genetic variation
Abstract
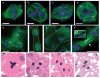
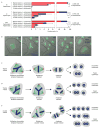
Mononucleated and binucleated polyploid hepatocytes (4n, 8n, 16n and higher) are found in all mammalian species, but the functional significance of this conserved phenomenon remains unknown. Polyploidization occurs through failed cytokinesis, begins at weaning in rodents and increases with age. Previously, we demonstrated that the opposite event, ploidy reversal, also occurs in polyploid hepatocytes generated by artificial cell fusion. This raised the possibility that somatic 'reductive mitoses' can also happen in normal hepatocytes. Here we show that multipolar mitotic spindles form frequently in mouse polyploid hepatocytes and can result in one-step ploidy reversal to generate offspring with halved chromosome content. Proliferating hepatocytes produce a highly diverse population of daughter cells with multiple numerical chromosome imbalances as well as uniparental origins. Our findings support a dynamic model of hepatocyte polyploidization, ploidy reversal and aneuploidy, a phenomenon that we term the 'ploidy conveyor'. We propose that this mechanism evolved to generate genetic diversity and permits adaptation of hepatocytes to xenobiotic or nutritional injury.
Figures


Comment in
-
Liver: new insight into hepatocyte genetic diversity and adaptation to injury.Nat Rev Gastroenterol Hepatol. 2010 Dec;7(12):649. doi: 10.1038/nrgastro.2010.175. Nat Rev Gastroenterol Hepatol. 2010. PMID: 21171209 No abstract available.
Similar articles
-
Aneuploidy, polyploidy and ploidy reversal in the liver.Semin Cell Dev Biol. 2013 Apr;24(4):347-56. doi: 10.1016/j.semcdb.2013.01.003. Epub 2013 Jan 16. Semin Cell Dev Biol. 2013. PMID: 23333793 Review.
-
Frequent aneuploidy among normal human hepatocytes.Gastroenterology. 2012 Jan;142(1):25-8. doi: 10.1053/j.gastro.2011.10.029. Epub 2011 Nov 2. Gastroenterology. 2012. PMID: 22057114 Free PMC article.
-
In Vivo Lineage Tracing of Polyploid Hepatocytes Reveals Extensive Proliferation during Liver Regeneration.Cell Stem Cell. 2020 Jan 2;26(1):34-47.e3. doi: 10.1016/j.stem.2019.11.014. Epub 2019 Dec 19. Cell Stem Cell. 2020. PMID: 31866222 Free PMC article.
-
Proliferative polyploid cells give rise to tumors via ploidy reduction.Nat Commun. 2021 Jan 28;12(1):646. doi: 10.1038/s41467-021-20916-y. Nat Commun. 2021. PMID: 33510149 Free PMC article.
-
Regulation of cytokinesis and its clinical significance.Crit Rev Clin Lab Sci. 2015;52(4):159-67. doi: 10.3109/10408363.2015.1012191. Epub 2015 Jun 24. Crit Rev Clin Lab Sci. 2015. PMID: 26104038 Review.
Cited by
-
Bursty gene expression in the intact mammalian liver.Mol Cell. 2015 Apr 2;58(1):147-56. doi: 10.1016/j.molcel.2015.01.027. Epub 2015 Feb 26. Mol Cell. 2015. PMID: 25728770 Free PMC article.
-
Underlying potential: cellular and molecular determinants of adult liver repair.J Clin Invest. 2013 May;123(5):1858-60. doi: 10.1172/JCI69966. Epub 2013 May 1. J Clin Invest. 2013. PMID: 23635782 Free PMC article. Review.
-
Transient endoreplication down-regulates the kinesin-14 HSET and contributes to genomic instability.Mol Biol Cell. 2016 Oct 1;27(19):2911-23. doi: 10.1091/mbc.E16-03-0159. Epub 2016 Aug 3. Mol Biol Cell. 2016. PMID: 27489338 Free PMC article.
-
Ploidy dynamics increase the risk of liver cancer initiation.Nat Commun. 2021 Mar 25;12(1):1896. doi: 10.1038/s41467-021-21897-8. Nat Commun. 2021. PMID: 33767143 Free PMC article.
-
Cellular origins of regenerating liver and hepatocellular carcinoma.JHEP Rep. 2021 Dec 13;4(4):100416. doi: 10.1016/j.jhepr.2021.100416. eCollection 2022 Apr. JHEP Rep. 2021. PMID: 35243280 Free PMC article. Review.
References
-
- Faktor VM, Uryvaeva IV. Progressive polyploidy in mouse liver following repeated hepatectomy. Tsitologiia. 1975;17:909–16. - PubMed
-
- Guidotti JE, et al. Liver cell polyploidization: a pivotal role for binuclear hepatocytes. J Biol Chem. 2003;278:19095–101. - PubMed
-
- Kudryavtsev BN, Kudryavtseva MV, Sakuta GA, Stein GI. Human hepatocyte polyploidization kinetics in the course of life cycle. Virchows Arch B Cell Pathol Incl Mol Pathol. 1993;64:387–93. - PubMed
-
- Barbason H, Van Cantfort J, Houbrechts N. Correlation between tissular and division functions in the liver of young rats. Cell Tissue Kinet. 1974;7:319–26. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

