DNA is a co-factor for its own replication in Xenopus egg extracts
- PMID: 20861001
- PMCID: PMC3025559
- DOI: 10.1093/nar/gkq739
DNA is a co-factor for its own replication in Xenopus egg extracts
Abstract
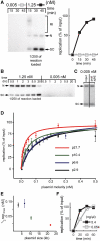
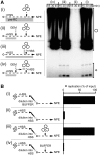
Soluble Xenopus egg extracts efficiently replicate added plasmids using a physiological mechanism, and thus represent a powerful system to understand vertebrate DNA replication. Surprisingly, DNA replication in this system is highly sensitive to plasmid concentration, being undetectable below ∼10 pM and highly efficient above ∼75 pM. DNA replication at the high plasmid concentration does not require plasmid-plasmid contacts, since replication is not inhibited when plasmids are immobilized in agarose prior to addition of egg extract. The absence of replication at low plasmid concentration is due to a defect in the assembly of pre-replication complexes (pre-RCs). pre-RC assembly requires contact-independent communication between plasmids. Our results show that in Xenopus egg extracts, aggregation of multiple replication forks is not required for efficient replication of plasmid DNA, and they suggest that DNA functions as a co-factor for its own duplication.
Figures


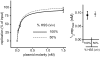

Similar articles
-
De novo assembly of genuine replication forks on an immobilized circular plasmid in Xenopus egg extracts.Nucleic Acids Res. 2006 Jul 26;34(13):e91. doi: 10.1093/nar/gkl512. Nucleic Acids Res. 2006. PMID: 16870720 Free PMC article.
-
Efficient plasmid DNA replication in Xenopus egg extracts does not depend on prior chromatin assembly.J Biol Chem. 1995 Dec 15;270(50):29676-81. doi: 10.1074/jbc.270.50.29676. J Biol Chem. 1995. PMID: 8530355
-
DNA replication initiates at multiple sites on plasmid DNA in Xenopus egg extracts.Nucleic Acids Res. 1992 Apr 11;20(7):1457-62. doi: 10.1093/nar/20.7.1457. Nucleic Acids Res. 1992. PMID: 1579437 Free PMC article.
-
Chromosomal DNA replication in a soluble cell-free system derived from Xenopus eggs.Methods Mol Biol. 2006;322:121-37. doi: 10.1007/978-1-59745-000-3_9. Methods Mol Biol. 2006. PMID: 16739720 Review.
-
Origin-specific initiation of mammalian nuclear DNA replication in a Xenopus cell-free system.Methods. 1997 Nov;13(3):313-24. doi: 10.1006/meth.1997.0530. Methods. 1997. PMID: 9441857 Review.
Cited by
-
Single-molecule analysis of DNA replication in Xenopus egg extracts.Methods. 2012 Jun;57(2):179-86. doi: 10.1016/j.ymeth.2012.03.033. Epub 2012 Apr 6. Methods. 2012. PMID: 22503776 Free PMC article. Review.
-
FANCJ promotes DNA synthesis through G-quadruplex structures.EMBO J. 2014 Nov 3;33(21):2521-33. doi: 10.15252/embj.201488663. Epub 2014 Sep 5. EMBO J. 2014. PMID: 25193968 Free PMC article.
-
Ensemble and Single-Molecule Analysis of Non-Homologous End Joining in Frog Egg Extracts.Methods Enzymol. 2017;591:233-270. doi: 10.1016/bs.mie.2017.03.020. Epub 2017 May 15. Methods Enzymol. 2017. PMID: 28645371 Free PMC article.
-
DNA replication origin activation in space and time.Nat Rev Mol Cell Biol. 2015 Jun;16(6):360-74. doi: 10.1038/nrm4002. Nat Rev Mol Cell Biol. 2015. PMID: 25999062 Review.
-
XLF acts as a flexible connector during non-homologous end joining.Elife. 2020 Dec 8;9:e61920. doi: 10.7554/eLife.61920. Elife. 2020. PMID: 33289484 Free PMC article.
References
-
- Takeda DY, Dutta A. DNA replication and progression through S phase. Oncogene. 2005;24:2827–2843. - PubMed
-
- Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002;71:333–374. - PubMed
-
- Arias EE, Walter JC. Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev. 2007;21:497–518. - PubMed
-
- Diffley JF. DNA replication: building the perfect switch. Curr. Biol. 2001;11:R367–R370. - PubMed

