The stepping pattern of myosin X is adapted for processive motility on bundled actin
- PMID: 20858426
- PMCID: PMC2941030
- DOI: 10.1016/j.bpj.2010.06.066
The stepping pattern of myosin X is adapted for processive motility on bundled actin
Abstract
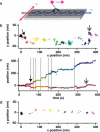
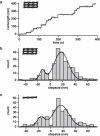
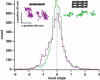
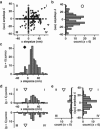
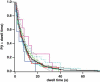
Myosin X is a molecular motor that is adapted to select bundled actin filaments over single actin filaments for processive motility. Its unique form of motility suggests that myosin X's stepping mechanism takes advantage of the arrangement of actin filaments and the additional target binding sites found within a bundle. Here we use fluorescence imaging with one-nanometer accuracy to show that myosin X takes steps of ∼18 nm along a fascin-actin bundle. This step-size is well short of the 36-nm step-size observed in myosin V and myosin VI that corresponds to the actin pseudohelical repeat distance. Myosin X is able to walk along bundles with this step-size if it straddles two actin filaments, but would be quickly forced to spiral into the constrained interior of the bundle if it were to use only a single actin filament. We also demonstrate that myosin X takes many sideways steps as it walks along a bundle, suggesting that it can switch actin filament pairs within the bundle as it walks. Sideways steps to the left or the right occur on bundles with equal frequency, suggesting a degree of lateral flexibility such that the motor's working stroke does not bias it to the left or to the right. On single actin filaments, we find a broad mixture of 10-20-nm steps, which again falls short of the 36-nm actin repeat. Moreover, the motor leans to the right as it walks along single filaments, which may require myosin X to adopt strained configurations. As a control, we also tracked myosin V stepping along actin filaments and fascin-actin bundles. We find that myosin V follows a narrower path on both structures, walking primarily along one surface of an actin filament and following a single filament within a bundle while occasionally switching to neighboring filaments. Together, these results delineate some of the structural features of the motor and the track that allow myosin X to recognize actin filament bundles.
Copyright © 2010 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
Similar articles
-
Actin structure-dependent stepping of myosin 5a and 10 during processive movement.PLoS One. 2013 Sep 19;8(9):e74936. doi: 10.1371/journal.pone.0074936. eCollection 2013. PLoS One. 2013. PMID: 24069366 Free PMC article.
-
A myosin motor that selects bundled actin for motility.Proc Natl Acad Sci U S A. 2008 Jul 15;105(28):9616-20. doi: 10.1073/pnas.0802592105. Epub 2008 Jul 3. Proc Natl Acad Sci U S A. 2008. PMID: 18599451 Free PMC article.
-
The Antiparallel Dimerization of Myosin X Imparts Bundle Selectivity for Processive Motility.Biophys J. 2018 Mar 27;114(6):1400-1410. doi: 10.1016/j.bpj.2018.01.038. Biophys J. 2018. PMID: 29590597 Free PMC article.
-
The complexity and diversity of the actin cytoskeleton of trypanosomatids.Mol Biochem Parasitol. 2020 May;237:111278. doi: 10.1016/j.molbiopara.2020.111278. Epub 2020 Apr 28. Mol Biochem Parasitol. 2020. PMID: 32353561 Review.
-
Biochemistry of Drebrin and Its Binding to Actin Filaments.Adv Exp Med Biol. 2017;1006:37-47. doi: 10.1007/978-4-431-56550-5_3. Adv Exp Med Biol. 2017. PMID: 28865013 Review.
Cited by
-
Actin binding proteins: their ups and downs in metastatic life.Cell Adh Migr. 2013 Mar-Apr;7(2):199-213. doi: 10.4161/cam.23176. Epub 2013 Jan 9. Cell Adh Migr. 2013. PMID: 23302954 Free PMC article. Review.
-
The ATPase mechanism of myosin 15, the molecular motor mutated in DFNB3 human deafness.J Biol Chem. 2021 Jan-Jun;296:100243. doi: 10.1074/jbc.RA120.014903. Epub 2021 Jan 9. J Biol Chem. 2021. PMID: 33372036 Free PMC article.
-
Use of fluorescent techniques to study the in vitro movement of myosins.Exp Suppl. 2014;105:193-210. doi: 10.1007/978-3-0348-0856-9_9. Exp Suppl. 2014. PMID: 25095996 Free PMC article. Review.
-
Myosin-X and disease.Exp Cell Res. 2015 May 15;334(1):10-5. doi: 10.1016/j.yexcr.2015.03.014. Epub 2015 Mar 27. Exp Cell Res. 2015. PMID: 25819274 Free PMC article. Review.
-
Future challenges in single-molecule fluorescence and laser trap approaches to studies of molecular motors.Dev Cell. 2012 Dec 11;23(6):1084-91. doi: 10.1016/j.devcel.2012.10.002. Dev Cell. 2012. PMID: 23237942 Free PMC article. Review.
References
-
- Berg J.S., Cheney R.E. Myosin-X is an unconventional myosin that undergoes intrafilopodial motility. Nat. Cell Biol. 2002;4:246–250. - PubMed
-
- Berg J.S., Derfler B.H., Cheney R.E. Myosin-X, a novel myosin with pleckstrin homology domains, associates with regions of dynamic actin. J. Cell Sci. 2000;113:3439–3451. - PubMed
-
- Zhang H., Berg J.S., Strömblad S. Myosin-X provides a motor-based link between integrins and the cytoskeleton. Nat. Cell Biol. 2004;6:523–531. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

