Transcriptional repressor E4-binding protein 4 (E4BP4) regulates metabolic hormone fibroblast growth factor 21 (FGF21) during circadian cycles and feeding
- PMID: 20851878
- PMCID: PMC2978569
- DOI: 10.1074/jbc.M110.172866
Transcriptional repressor E4-binding protein 4 (E4BP4) regulates metabolic hormone fibroblast growth factor 21 (FGF21) during circadian cycles and feeding
Abstract
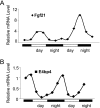
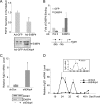
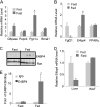
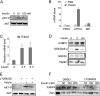
Fibroblast growth factor 21 (FGF21) is a potent antidiabetic and triglyceride-lowering hormone whose hepatic expression is highly responsive to food intake. FGF21 induction in the adaptive response to fasting has been well studied, but the molecular mechanism responsible for feeding-induced repression remains unknown. In this study, we demonstrate a novel link between FGF21 and a key circadian output protein, E4BP4. Expression of Fgf21 displays a circadian rhythm, which peaks during the fasting phase and is anti-phase to E4bp4, which is elevated during feeding periods. E4BP4 strongly suppresses Fgf21 transcription by binding to a D-box element in the distal promoter region. Depletion of E4BP4 in synchronized Hepa1c1c-7 liver cells augments the amplitude of Fgf21 expression, and overexpression of E4BP4 represses FGF21 secretion from primary mouse hepatocytes. Mimicking feeding effects, insulin significantly increases E4BP4 expression and binding to the Fgf21 promoter through AKT activation. Thus, E4BP4 is a novel insulin-responsive repressor of FGF21 expression during circadian cycles and feeding.
Figures

Similar articles
-
Recruitment of histone methyltransferase G9a mediates transcriptional repression of Fgf21 gene by E4BP4 protein.J Biol Chem. 2013 Feb 22;288(8):5417-25. doi: 10.1074/jbc.M112.433482. Epub 2013 Jan 2. J Biol Chem. 2013. PMID: 23283977 Free PMC article.
-
Clock-controlled output gene Dbp is a regulator of Arnt/Hif-1β gene expression in pancreatic islet β-cells.Biochem Biophys Res Commun. 2013 May 3;434(2):370-5. doi: 10.1016/j.bbrc.2013.03.084. Epub 2013 Apr 6. Biochem Biophys Res Commun. 2013. PMID: 23567972
-
A novel E4BP4 element drives circadian expression of mPeriod2.Nucleic Acids Res. 2007;35(2):648-55. doi: 10.1093/nar/gkl868. Epub 2006 Dec 19. Nucleic Acids Res. 2007. PMID: 17182630 Free PMC article.
-
Hepatic metabolic regulation by nuclear factor E4BP4.J Mol Endocrinol. 2021 Jan;66(1):R15-R21. doi: 10.1530/JME-20-0239. J Mol Endocrinol. 2021. PMID: 33434146 Free PMC article. Review.
-
The regulation of FGF21 gene expression by metabolic factors and nutrients.Horm Mol Biol Clin Investig. 2016 Jun 10;30(1):/j/hmbci.2017.30.issue-1/hmbci-2016-0016/hmbci-2016-0016.xml. doi: 10.1515/hmbci-2016-0016. Horm Mol Biol Clin Investig. 2016. PMID: 27285327 Review.
Cited by
-
Hepatic E4BP4 induction promotes lipid accumulation by suppressing AMPK signaling in response to chemical or diet-induced ER stress.FASEB J. 2020 Oct;34(10):13533-13547. doi: 10.1096/fj.201903292RR. Epub 2020 Aug 11. FASEB J. 2020. PMID: 32780887 Free PMC article.
-
Recruitment of histone methyltransferase G9a mediates transcriptional repression of Fgf21 gene by E4BP4 protein.J Biol Chem. 2013 Feb 22;288(8):5417-25. doi: 10.1074/jbc.M112.433482. Epub 2013 Jan 2. J Biol Chem. 2013. PMID: 23283977 Free PMC article.
-
Disturbances in the murine hepatic circadian clock in alcohol-induced hepatic steatosis.Sci Rep. 2014 Jan 16;4:3725. doi: 10.1038/srep03725. Sci Rep. 2014. PMID: 24430730 Free PMC article.
-
The Nuclear Receptor Rev-erbα Regulates Adipose Tissue-specific FGF21 Signaling.J Biol Chem. 2016 May 13;291(20):10867-75. doi: 10.1074/jbc.M116.719120. Epub 2016 Mar 21. J Biol Chem. 2016. PMID: 27002153 Free PMC article.
-
Alterations in Hepatic FGF21, Co-Regulated Genes, and Upstream Metabolic Genes in Response to Nutrition, Ketosis and Inflammation in Peripartal Holstein Cows.PLoS One. 2015 Oct 9;10(10):e0139963. doi: 10.1371/journal.pone.0139963. eCollection 2015. PLoS One. 2015. PMID: 26451842 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

