Crystallographic and nuclear magnetic resonance evaluation of the impact of peptide binding to the second PDZ domain of protein tyrosine phosphatase 1E
- PMID: 20839809
- PMCID: PMC3001272
- DOI: 10.1021/bi101131f
Crystallographic and nuclear magnetic resonance evaluation of the impact of peptide binding to the second PDZ domain of protein tyrosine phosphatase 1E
Abstract
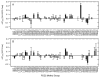
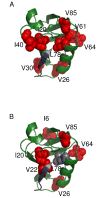
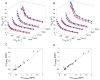
PDZ (PSD95/Discs large/ZO-1) domains are ubiquitous protein interaction motifs found in scaffolding proteins involved in signal transduction. Despite the fact that many PDZ domains show a limited tendency to undergo structural change, the PDZ family has been associated with long-range communication and allostery. One of the PDZ domains studied most in terms of structure and biophysical properties is the second PDZ ("PDZ2") domain from protein tyrosine phosphatase 1E (PTP1E, also known as PTPL1). Previously, we showed through NMR relaxation studies that binding of the RA-GEF2 C-terminal peptide substrate results in long-range propagation of side-chain dynamic changes in human PDZ2 [Fuentes, E. J., et al. (2004) J. Mol. Biol. 335, 1105-1115]. Here, we present the first X-ray crystal structures of PDZ2 in the absence and presence of RA-GEF2 ligand, determined to resolutions of 1.65 and 1.3 Å, respectively. These structures deviate somewhat from previously determined NMR structures and indicate that very minor structural changes in PDZ2 accompany peptide binding. NMR residual dipolar couplings confirm the crystal structures to be accurate models of the time-averaged atomic coordinates of PDZ2. The impact on side-chain dynamics was further tested with a C-terminal peptide from APC, which showed results nearly identical to those of RA-GEF2. Thus, allosteric transmission in PDZ2 induced by peptide binding is conveyed purely and robustly by dynamics. (15)N relaxation dispersion measurements did not detect appreciable populations of a kinetic structural intermediate. Collectively, for ligand binding to PDZ2, these data support a lock-and-key binding model from a structural perspective and an allosteric model from a dynamical perspective, which together suggest a complex energy landscape for functional transitions within the ensemble.
Figures
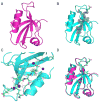
Similar articles
-
An allosteric intramolecular PDZ-PDZ interaction modulates PTP-BL PDZ2 binding specificity.Biochemistry. 2007 Nov 27;46(47):13629-37. doi: 10.1021/bi700954e. Epub 2007 Nov 3. Biochemistry. 2007. PMID: 17979300
-
The binding affinity of PTPN13's tandem PDZ2/3 domain is allosterically modulated.BMC Mol Cell Biol. 2019 Jul 8;20(1):23. doi: 10.1186/s12860-019-0203-6. BMC Mol Cell Biol. 2019. PMID: 31286859 Free PMC article.
-
Atomic resolution protein allostery from the multi-state structure of a PDZ domain.Nat Commun. 2022 Oct 20;13(1):6232. doi: 10.1038/s41467-022-33687-x. Nat Commun. 2022. PMID: 36266302 Free PMC article.
-
Change in allosteric network affects binding affinities of PDZ domains: analysis through perturbation response scanning.PLoS Comput Biol. 2011 Oct;7(10):e1002154. doi: 10.1371/journal.pcbi.1002154. Epub 2011 Oct 6. PLoS Comput Biol. 2011. PMID: 21998559 Free PMC article. Review.
-
Allosterism in the PDZ Family.Int J Mol Sci. 2022 Jan 27;23(3):1454. doi: 10.3390/ijms23031454. Int J Mol Sci. 2022. PMID: 35163402 Free PMC article. Review.
Cited by
-
Kinetic response of a photoperturbed allosteric protein.Proc Natl Acad Sci U S A. 2013 Jul 16;110(29):11725-30. doi: 10.1073/pnas.1306323110. Epub 2013 Jul 1. Proc Natl Acad Sci U S A. 2013. PMID: 23818626 Free PMC article.
-
Dynamic allostery: linkers are not merely flexible.Structure. 2011 Jul 13;19(7):907-17. doi: 10.1016/j.str.2011.06.002. Structure. 2011. PMID: 21742258 Free PMC article. Review.
-
The underappreciated role of allostery in the cellular network.Annu Rev Biophys. 2013;42:169-89. doi: 10.1146/annurev-biophys-083012-130257. Epub 2013 Feb 28. Annu Rev Biophys. 2013. PMID: 23451894 Free PMC article. Review.
-
Interaction between SARS-CoV PBM and Cellular PDZ Domains Leading to Virus Virulence.Viruses. 2024 Jul 29;16(8):1214. doi: 10.3390/v16081214. Viruses. 2024. PMID: 39205188 Free PMC article.
-
The spatial structure of cell signaling systems.Phys Biol. 2013 Aug;10(4):045004. doi: 10.1088/1478-3975/10/4/045004. Epub 2013 Aug 2. Phys Biol. 2013. PMID: 23913102 Free PMC article.
References
-
- Kim E, Sheng M. PDZ domain proteins of synapses. Nat Rev Neurosci. 2004;5:771–781. - PubMed
-
- Sheng M, Sala C. PDZ domains and the organization of supramolecular complexes. Annu Rev Neurosci. 2001;24:1–29. - PubMed
-
- van Ham M, Hendriks W. PDZ domains-glue and guide. Mol Biol Rep. 2003;30:69–82. - PubMed
-
- Peterson FC, Penkert RR, Volkman BF, Prehoda KE. Cdc42 regulates the Par-6 PDZ domain through an allosteric CRIB-PDZ transition. Mol Cell. 2004;13:665–676. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

