A dual E3 mechanism for Rub1 ligation to Cdc53
- PMID: 20832729
- PMCID: PMC3001161
- DOI: 10.1016/j.molcel.2010.08.030
A dual E3 mechanism for Rub1 ligation to Cdc53
Abstract
In ubiquitin-like protein (UBL) cascades, a thioester-linked E2∼UBL complex typically interacts with an E3 enzyme for UBL transfer to the target. Here we demonstrate a variant mechanism, whereby the E2 Ubc12 functions with two E3s, Hrt1 and Dcn1, for ligation of the UBL Rub1 to Cdc53's WHB subdomain. Hrt1 functions like a conventional RING E3, with its N terminus recruiting Cdc53 and C-terminal RING activating Ubc12∼Rub1. Dcn1's "potentiating neddylation" domain (Dcn1(P)) acts as an additional E3, reducing nonspecific Hrt1-mediated Ubc12∼Rub1 discharge and directing Ubc12's active site to Cdc53. Crystal structures of Dcn1(P)-Cdc53(WHB) and Ubc12 allow modeling of a catalytic complex, supported by mutational data. We propose that Dcn1's interactions with both Cdc53 and Ubc12 would restrict the otherwise flexible Hrt1 RING-bound Ubc12∼Rub1 to a catalytically competent orientation. Our data reveal mechanisms by which two E3s function synergistically to promote UBL transfer from one E2 to a target.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures



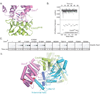
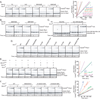

Similar articles
-
A RING E3-substrate complex poised for ubiquitin-like protein transfer: structural insights into cullin-RING ligases.Nat Struct Mol Biol. 2011 Jul 17;18(8):947-9. doi: 10.1038/nsmb.2086. Nat Struct Mol Biol. 2011. PMID: 21765416 Free PMC article.
-
A longevity protein, Lag2, interacts with SCF complex and regulates SCF function.EMBO J. 2009 Nov 4;28(21):3366-77. doi: 10.1038/emboj.2009.268. Epub 2009 Sep 17. EMBO J. 2009. PMID: 19763088 Free PMC article.
-
Structure of a RING E3 trapped in action reveals ligation mechanism for the ubiquitin-like protein NEDD8.Cell. 2014 Jun 19;157(7):1671-84. doi: 10.1016/j.cell.2014.04.037. Cell. 2014. PMID: 24949976 Free PMC article.
-
NEDD8 and ubiquitin ligation by cullin-RING E3 ligases.Curr Opin Struct Biol. 2021 Apr;67:101-109. doi: 10.1016/j.sbi.2020.10.007. Epub 2020 Nov 5. Curr Opin Struct Biol. 2021. PMID: 33160249 Free PMC article. Review.
-
Genetically engineered mouse models for functional studies of SKP1-CUL1-F-box-protein (SCF) E3 ubiquitin ligases.Cell Res. 2013 May;23(5):599-619. doi: 10.1038/cr.2013.44. Epub 2013 Mar 26. Cell Res. 2013. PMID: 23528706 Free PMC article. Review.
Cited by
-
Hypertension-causing Mutations in Cullin3 Protein Impair RhoA Protein Ubiquitination and Augment the Association with Substrate Adaptors.J Biol Chem. 2015 Jul 31;290(31):19208-17. doi: 10.1074/jbc.M115.645358. Epub 2015 Jun 22. J Biol Chem. 2015. PMID: 26100637 Free PMC article.
-
The N-end rule pathway is mediated by a complex of the RING-type Ubr1 and HECT-type Ufd4 ubiquitin ligases.Nat Cell Biol. 2010 Dec;12(12):1177-85. doi: 10.1038/ncb2121. Epub 2010 Nov 14. Nat Cell Biol. 2010. PMID: 21076411 Free PMC article.
-
A spectrophotometric assay for conjugation of ubiquitin and ubiquitin-like proteins.Anal Biochem. 2011 Nov 1;418(1):102-10. doi: 10.1016/j.ab.2011.06.034. Epub 2011 Jul 2. Anal Biochem. 2011. PMID: 21771579 Free PMC article.
-
Characterization of the mammalian family of DCN-type NEDD8 E3 ligases.J Cell Sci. 2016 Apr 1;129(7):1441-54. doi: 10.1242/jcs.181784. Epub 2016 Feb 18. J Cell Sci. 2016. PMID: 26906416 Free PMC article.
-
The Ubiquitin-like Proteins of Saccharomyces cerevisiae.Biomolecules. 2023 Apr 24;13(5):734. doi: 10.3390/biom13050734. Biomolecules. 2023. PMID: 37238603 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
- R01 GM069530-06/GM/NIGMS NIH HHS/United States
- P41 RR015301/RR/NCRR NIH HHS/United States
- R01 GM069530-07/GM/NIGMS NIH HHS/United States
- R01 GM077053/GM/NIGMS NIH HHS/United States
- RR-15301/RR/NCRR NIH HHS/United States
- R01CA082491/CA/NCI NIH HHS/United States
- R01 CA082491-08/CA/NCI NIH HHS/United States
- P30 CA021765/CA/NCI NIH HHS/United States
- P41 RR015301-075431/RR/NCRR NIH HHS/United States
- P30 CA021765-31/CA/NCI NIH HHS/United States
- R01 CA082491/CA/NCI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- P30 CA021765-32/CA/NCI NIH HHS/United States
- R01 GM077053-04/GM/NIGMS NIH HHS/United States
- R01 GM069530/GM/NIGMS NIH HHS/United States
- R01 GM069530-08/GM/NIGMS NIH HHS/United States
- R01GM069530/GM/NIGMS NIH HHS/United States
- R01 CA082491-09/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

