Structural studies and the assembly of the heptameric post-translational translocon complex
- PMID: 20826819
- PMCID: PMC3024790
- DOI: 10.1074/jbc.M110.159517
Structural studies and the assembly of the heptameric post-translational translocon complex
Abstract
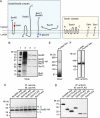
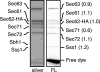
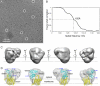
In Saccharomyces cerevisiae, some of the nascent chains can be post-translationally translocated into the endoplasmic reticulum through the heptameric post-translational translocon complex (post-translocon). This membrane-protein complex is composed of the protein-conducting channel and the tetrameric Sec62/63 complex. The Sec62/63 complex plays crucial roles in targeting of the signal recognition particle-independent protein substrate to the protein-conducting channel and in assembly of the post-translocon. Although the molecular mechanism of the post-translational translocation process has been well established, the structure of the post-translocon and how the channel and the Sec62/63 complex form the heptameric complex are largely uncharacterized. Here, we report a 20-Å resolution cryo-electron microscopy structure of the post-translocon. The purified post-translocon was found to have a mass of 287 kDa, which is consistent with the unit stoichiometry of the seven subunits as determined by a cysteine labeling experiment. We demonstrated that Triton X-100 dissociated the heptameric complex into three subcomplexes identified as the trimeric translocon Sec61-Sbh1-Sss1, the Sec63-Sec71-Sec72 trimer, and the heterotetramer Sec62-Sec63-Sec71-Sec72, respectively. Additionally, a role of the sixth cytosolic loop of Sec61 in assembly of the post-translocon was demonstrated. Mutations of conserved, positively charged amino acid residues in the loop caused decreased formation of the post-translocon. These studies provide the first architectural description of the yeast post-translocon.
Figures


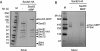


Similar articles
-
Emerging View on the Molecular Functions of Sec62 and Sec63 in Protein Translocation.Int J Mol Sci. 2021 Nov 25;22(23):12757. doi: 10.3390/ijms222312757. Int J Mol Sci. 2021. PMID: 34884562 Free PMC article. Review.
-
Structure of the post-translational protein translocation machinery of the ER membrane.Nature. 2019 Feb;566(7742):136-139. doi: 10.1038/s41586-018-0856-x. Epub 2018 Dec 31. Nature. 2019. PMID: 30644436 Free PMC article.
-
Architecture of the active post-translational Sec translocon.EMBO J. 2021 Feb 1;40(3):e105643. doi: 10.15252/embj.2020105643. Epub 2020 Dec 11. EMBO J. 2021. PMID: 33305433 Free PMC article.
-
Two alternative binding mechanisms connect the protein translocation Sec71-Sec72 complex with heat shock proteins.J Biol Chem. 2017 May 12;292(19):8007-8018. doi: 10.1074/jbc.M116.761122. Epub 2017 Mar 12. J Biol Chem. 2017. PMID: 28286332 Free PMC article.
-
A clearer picture of the ER translocon complex.J Cell Sci. 2020 Feb 4;133(3):jcs231340. doi: 10.1242/jcs.231340. J Cell Sci. 2020. PMID: 32019826 Review.
Cited by
-
The Protease Ste24 Clears Clogged Translocons.Cell. 2016 Jan 14;164(1-2):103-114. doi: 10.1016/j.cell.2015.11.053. Cell. 2016. PMID: 26771486 Free PMC article.
-
An EGF-like protein forms a complex with PfRh5 and is required for invasion of human erythrocytes by Plasmodium falciparum.PLoS Pathog. 2011 Sep;7(9):e1002199. doi: 10.1371/journal.ppat.1002199. Epub 2011 Sep 1. PLoS Pathog. 2011. PMID: 21909261 Free PMC article.
-
The SND proteins constitute an alternative targeting route to the endoplasmic reticulum.Nature. 2016 Nov 30;540(7631):134-138. doi: 10.1038/nature20169. Nature. 2016. PMID: 27905431 Free PMC article.
-
Protein translocation across the rough endoplasmic reticulum.Cold Spring Harb Perspect Biol. 2013 Feb 1;5(2):a013342. doi: 10.1101/cshperspect.a013342. Cold Spring Harb Perspect Biol. 2013. PMID: 23251026 Free PMC article. Review.
-
The Sec63p J-domain is required for ERAD of soluble proteins in yeast.PLoS One. 2013 Dec 4;8(12):e82058. doi: 10.1371/journal.pone.0082058. eCollection 2013. PLoS One. 2013. PMID: 24324744 Free PMC article.
References
-
- Rapoport T. A. (2007) Nature 450, 663–669 - PubMed
-
- Stroud R. M., Walter P. (1999) Curr. Opin. Struct. Biol. 9, 754–759 - PubMed
-
- Wilkinson B. M., Critchley A. J., Stirling C. J. (1996) J. Biol. Chem. 271, 25590–25597 - PubMed
-
- Van den Berg B., Clemons W. M., Jr., Collinson I., Modis Y., Hartmann E., Harrison S. C., Rapoport T. A. (2004) Nature 427, 36–44 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

