tRNA biology charges to the front
- PMID: 20810645
- PMCID: PMC2932967
- DOI: 10.1101/gad.1956510
tRNA biology charges to the front
Abstract
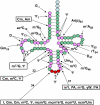
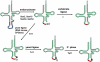
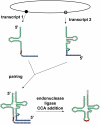
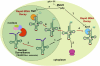
tRNA biology has come of age, revealing an unprecedented level of understanding and many unexpected discoveries along the way. This review highlights new findings on the diverse pathways of tRNA maturation, and on the formation and function of a number of modifications. Topics of special focus include the regulation of tRNA biosynthesis, quality control tRNA turnover mechanisms, widespread tRNA cleavage pathways activated in response to stress and other growth conditions, emerging evidence of signaling pathways involving tRNA and cleavage fragments, and the sophisticated intracellular tRNA trafficking that occurs during and after biosynthesis.
Figures
Similar articles
-
The life and times of a tRNA.RNA. 2023 Jul;29(7):898-957. doi: 10.1261/rna.079620.123. Epub 2023 Apr 13. RNA. 2023. PMID: 37055150 Free PMC article. Review.
-
Transfer RNAs: diversity in form and function.RNA Biol. 2021 Mar;18(3):316-339. doi: 10.1080/15476286.2020.1809197. Epub 2020 Sep 9. RNA Biol. 2021. PMID: 32900285 Free PMC article. Review.
-
An interplay between transcription, processing, and degradation determines tRNA levels in yeast.Wiley Interdiscip Rev RNA. 2013 Nov-Dec;4(6):709-22. doi: 10.1002/wrna.1190. Epub 2013 Aug 23. Wiley Interdiscip Rev RNA. 2013. PMID: 24039171 Review.
-
Quality Control Pathways for Nucleus-Encoded Eukaryotic tRNA Biosynthesis and Subcellular Trafficking.Mol Cell Biol. 2015 Jun;35(12):2052-8. doi: 10.1128/MCB.00131-15. Epub 2015 Apr 6. Mol Cell Biol. 2015. PMID: 25848089 Free PMC article. Review.
-
Cellular dynamics of tRNAs and their genes.FEBS Lett. 2010 Jan 21;584(2):310-7. doi: 10.1016/j.febslet.2009.11.053. FEBS Lett. 2010. PMID: 19931532 Free PMC article. Review.
Cited by
-
Identification of determinants for tRNA substrate recognition by Escherichia coli C/U34 2'-O-methyltransferase.RNA Biol. 2015;12(8):900-11. doi: 10.1080/15476286.2015.1050576. RNA Biol. 2015. PMID: 26106808 Free PMC article.
-
tRNA-derived small RNAs: Mechanisms and potential roles in cancers.Genes Dis. 2022 Jan 10;9(6):1431-1442. doi: 10.1016/j.gendis.2021.12.009. eCollection 2022 Nov. Genes Dis. 2022. PMID: 36157501 Free PMC article. Review.
-
Roles of Fe-S proteins: from cofactor synthesis to iron homeostasis to protein synthesis.Curr Opin Genet Dev. 2016 Jun;38:45-51. doi: 10.1016/j.gde.2016.03.006. Epub 2016 Apr 7. Curr Opin Genet Dev. 2016. PMID: 27061491 Free PMC article. Review.
-
The fitness landscape of a tRNA gene.Science. 2016 May 13;352(6287):837-40. doi: 10.1126/science.aae0568. Epub 2016 Apr 14. Science. 2016. PMID: 27080104 Free PMC article.
-
tRNA-modifying enzyme mutations induce codon-specific mistranslation and protein aggregation in yeast.RNA Biol. 2021 Apr;18(4):563-575. doi: 10.1080/15476286.2020.1819671. Epub 2020 Sep 17. RNA Biol. 2021. PMID: 32893724 Free PMC article.
References
-
- Acquati F, Possati L, Ferrante L, Campomenosi P, Talevi S, Bardelli S, Margiotta C, Russo A, Bortoletto E, Rocchetti R, et al. 2005. Tumor and metastasis suppression by the human RNASET2 gene. Int J Oncol 26: 1159–1168 - PubMed
-
- Aebi M, Kirchner G, Chen JY, Vijayraghavan U, Jacobson A, Martin NC, Abelson J 1990. Isolation of a temperature-sensitive mutant with an altered tRNA nucleotidyltransferase and cloning of the gene encoding tRNA nucleotidyltransferase in the yeast Saccharomyces cerevisiae. J Biol Chem 265: 16216–16220 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
