Gene set analysis exploiting the topology of a pathway
- PMID: 20809931
- PMCID: PMC2945950
- DOI: 10.1186/1752-0509-4-121
Gene set analysis exploiting the topology of a pathway
Abstract
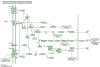
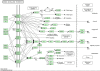
Background: Recently, a great effort in microarray data analysis is directed towards the study of the so-called gene sets. A gene set is defined by genes that are, somehow, functionally related. For example, genes appearing in a known biological pathway naturally define a gene set. The gene sets are usually identified from a priori biological knowledge. Nowadays, many bioinformatics resources store such kind of knowledge (see, for example, the Kyoto Encyclopedia of Genes and Genomes, among others). Although pathways maps carry important information about the structure of correlation among genes that should not be neglected, the currently available multivariate methods for gene set analysis do not fully exploit it.
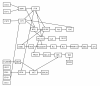
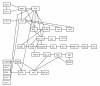
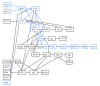
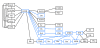
Results: We propose a novel gene set analysis specifically designed for gene sets defined by pathways. Such analysis, based on graphical models, explicitly incorporates the dependence structure among genes highlighted by the topology of pathways. The analysis is designed to be used for overall surveillance of changes in a pathway in different experimental conditions. In fact, under different circumstances, not only the expression of the genes in a pathway, but also the strength of their relations may change. The methods resulting from the proposal allow both to test for variations in the strength of the links, and to properly account for heteroschedasticity in the usual tests for differential expression.
Conclusions: The use of graphical models allows a deeper look at the components of the pathway that can be tested separately and compared marginally. In this way it is possible to test single components of the pathway and highlight only those involved in its deregulation.
Figures
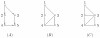

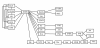
Similar articles
-
Path2enet: generation of human pathway-derived networks in an expression specific context.BMC Genomics. 2016 Oct 25;17(Suppl 8):731. doi: 10.1186/s12864-016-3066-7. BMC Genomics. 2016. PMID: 27801297 Free PMC article.
-
Probabilistic prioritization of candidate pathway association with pathway score.BMC Bioinformatics. 2018 Oct 24;19(1):391. doi: 10.1186/s12859-018-2411-z. BMC Bioinformatics. 2018. PMID: 30355338 Free PMC article.
-
Bi-directional gene set enrichment and canonical correlation analysis identify key diet-sensitive pathways and biomarkers of metabolic syndrome.BMC Bioinformatics. 2010 Oct 7;11:499. doi: 10.1186/1471-2105-11-499. BMC Bioinformatics. 2010. PMID: 20929581 Free PMC article.
-
Comparative study of gene set enrichment methods.BMC Bioinformatics. 2009 Sep 2;10:275. doi: 10.1186/1471-2105-10-275. BMC Bioinformatics. 2009. PMID: 19725948 Free PMC article.
-
An Experimental Approach to Genome Annotation: This report is based on a colloquium sponsored by the American Academy of Microbiology held July 19-20, 2004, in Washington, DC.Washington (DC): American Society for Microbiology; 2004. Washington (DC): American Society for Microbiology; 2004. PMID: 33001599 Free Books & Documents. Review.
Cited by
-
IPAVS: Integrated Pathway Resources, Analysis and Visualization System.Nucleic Acids Res. 2012 Jan;40(Database issue):D803-8. doi: 10.1093/nar/gkr1208. Epub 2011 Dec 2. Nucleic Acids Res. 2012. PMID: 22140115 Free PMC article.
-
Down-weighting overlapping genes improves gene set analysis.BMC Bioinformatics. 2012 Jun 19;13:136. doi: 10.1186/1471-2105-13-136. BMC Bioinformatics. 2012. PMID: 22713124 Free PMC article.
-
Graphite Web: Web tool for gene set analysis exploiting pathway topology.Nucleic Acids Res. 2013 Jul;41(Web Server issue):W89-97. doi: 10.1093/nar/gkt386. Epub 2013 May 10. Nucleic Acids Res. 2013. PMID: 23666626 Free PMC article.
-
graphite - a Bioconductor package to convert pathway topology to gene network.BMC Bioinformatics. 2012 Jan 31;13:20. doi: 10.1186/1471-2105-13-20. BMC Bioinformatics. 2012. PMID: 22292714 Free PMC article.
-
A comprehensive survey of the approaches for pathway analysis using multi-omics data integration.Brief Bioinform. 2022 Nov 19;23(6):bbac435. doi: 10.1093/bib/bbac435. Brief Bioinform. 2022. PMID: 36252928 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

