Induced Sézary syndrome PBMCs poorly express immune response genes up-regulated in stimulated memory T cells
- PMID: 20801618
- PMCID: PMC3928076
- DOI: 10.1016/j.jdermsci.2010.07.007
Induced Sézary syndrome PBMCs poorly express immune response genes up-regulated in stimulated memory T cells
Abstract
Background: Dysfunctions in memory T cells contribute to various inflammatory autoimmune diseases and neoplasms. We hypothesize that investigating the differences of genetic profiles between resting and activated naïve and memory T cells may provide insight into the characterization of abnormal memory T cells in diseases, such as Sézary syndrome (SS), a neoplasm composed of CD4(+) CD45RO(+) cells.
Objective: We determined genes distinctively expressed between resting and activated naive and memory cells. Levels of up-regulated genes in resting and activated memory cells were measured in SS PBMCs, which were largely comprised of CD4(+) CD45RO(+) cells, to quantitatively assess how different Sézary cells were from memory cells.
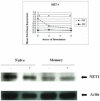
Methods: We compared gene expression profiles using high-density oligo-microarrays between resting and activated naïve and memory CD4(+) T cells. Differentially expressed genes were confirmed by qRT-PCR and immunoblotting. Levels of genes up-regulated in activated and resting memory T cells were determined in SS PBMCs by qRT-PCR.
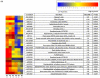
Results: Activated memory cells expressed greater numbers of immune-mediated genes involved in effector function compared to naïve cells in our microarray analysis and qRT-PCR. Nine out of 14 genes with enhanced levels in activated memory cells had reduced levels in SS PBMCs (p<0.05).
Conclusions: Activation of memory and naïve CD4(+) T cells revealed a diverging gap in gene expression between these subsets, with memory cells expressing immune-related genes important for effector function. Many of these genes were markedly depressed in SS patients, implying Sézary cells are markedly impaired in mounting immune responses compared to memory cells.
Copyright © 2010 Japanese Society for Investigative Dermatology. Published by Elsevier Ireland Ltd. All rights reserved.
Figures









Similar articles
-
Naïve/memory T-cell phenotypes in leukemic cutaneous T-cell lymphoma: Putative cell of origin overlaps disease classification.Cytometry B Clin Cytom. 2019 May;96(3):234-241. doi: 10.1002/cyto.b.21738. Epub 2018 Oct 16. Cytometry B Clin Cytom. 2019. PMID: 30328260 Free PMC article.
-
CD164 identifies CD4+ T cells highly expressing genes associated with malignancy in Sézary syndrome: the Sézary signature genes, FCRL3, Tox, and miR-214.Arch Dermatol Res. 2017 Jan;309(1):11-19. doi: 10.1007/s00403-016-1698-8. Epub 2016 Oct 20. Arch Dermatol Res. 2017. PMID: 27766406 Free PMC article.
-
Novel approach to gene expression profiling in Sézary syndrome.Br J Dermatol. 2010 Nov;163(5):1090-4. doi: 10.1111/j.1365-2133.2010.09973.x. Br J Dermatol. 2010. PMID: 20698843 Free PMC article.
-
Augmentation in expression of activation-induced genes differentiates memory from naive CD4+ T cells and is a molecular mechanism for enhanced cellular response of memory CD4+ T cells.J Immunol. 2001 Jun 15;166(12):7335-44. doi: 10.4049/jimmunol.166.12.7335. J Immunol. 2001. PMID: 11390484
-
Sézary syndrome: old enigmas, new targets.J Dtsch Dermatol Ges. 2016 Mar;14(3):256-64. doi: 10.1111/ddg.12900. J Dtsch Dermatol Ges. 2016. PMID: 26972187 Review.
Cited by
-
Gene Expression Comparison between Sézary Syndrome and Lymphocytic-Variant Hypereosinophilic Syndrome Refines Biomarkers for Sézary Syndrome.Cells. 2020 Aug 29;9(9):1992. doi: 10.3390/cells9091992. Cells. 2020. PMID: 32872487 Free PMC article. Review.
-
Evolving insights in the pathogenesis and therapy of cutaneous T-cell lymphoma (mycosis fungoides and Sezary syndrome).Br J Haematol. 2011 Oct;155(2):150-66. doi: 10.1111/j.1365-2141.2011.08852.x. Epub 2011 Aug 25. Br J Haematol. 2011. PMID: 21883142 Free PMC article. Review.
-
Transcriptome analysis of Sézary syndrome and lymphocytic-variant hypereosinophilic syndrome T cells reveals common and divergent genes.Oncotarget. 2019 Aug 20;10(49):5052-5069. doi: 10.18632/oncotarget.27120. eCollection 2019 Aug 20. Oncotarget. 2019. PMID: 31489115 Free PMC article.
References
-
- Luqman M, Bottomly K. Activation requirements for CD4+ T cells differing in CD45R expression. J Immunol. 1992;149:2300–6. - PubMed
-
- Tanchot C, Lemonnier FA, Perarnau B, Freitas AA, Rocha B. Differential requirements for survival and proliferation of CD8 naive or memory T cells. Science. 1997;276:2057–62. - PubMed
-
- London CA, Lodge MP, Abbas AK. Functional responses and costimulator dependence of memory CD4+ T cells. J Immunol. 2000;164:265–72. - PubMed
-
- Croft M, Bradley LM, Swain SL. Naive versus memory CD4 T cell response to antigen. Memory cells are less dependent on accessory cell costimulation and can respond to many antigen-presenting cell types including resting B cells. J Immunol. 1994;152:2675–85. - PubMed
-
- Van de Velde H, Lorre K, Bakkus M, Thielemans K, Ceuppens JL, de Boer M. CD45RO+ memory T cells but not CD45RA+ naive T cells can be efficiently activated by remote co-stimulation with B7. Int Immunol. 1993;5:1483–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

