A strategy for interaction site prediction between phospho-binding modules and their partners identified from proteomic data
- PMID: 20733106
- PMCID: PMC3101860
- DOI: 10.1074/mcp.M110.003319
A strategy for interaction site prediction between phospho-binding modules and their partners identified from proteomic data
Abstract
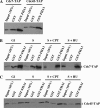
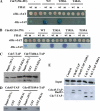
Small and large scale proteomic technologies are providing a wealth of potential interactions between proteins bearing phospho-recognition modules and their substrates. Resulting interaction maps reveal such a dense network of interactions that the functional dissection and understanding of these networks often require to break specific interactions while keeping the rest intact. Here, we developed a computational strategy, called STRIP, to predict the precise interaction site involved in an interaction with a phospho-recognition module. The method was validated by a two-hybrid screen carried out using the ForkHead Associated (FHA)1 domain of Rad53, a key protein of Saccharomyces cerevisiae DNA checkpoint, as a bait. In this screen we detected 11 partners, including Cdc7 and Cdc45, essential components of the DNA replication machinery. FHA domains are phospho-threonine binding modules and the threonines involved in both interactions could be predicted using the STRIP strategy. The threonines T484 and T189 in Cdc7 and Cdc45, respectively, were mutated and loss of binding could be monitored experimentally with the full-length proteins. The method was further tested for the analysis of 63 known Rad53 binding partners and provided several key insights regarding the threonines likely involved in these interactions. The STRIP method relies on a combination of conservation, phosphorylation likelihood, and binding specificity criteria and can be accessed via a web interface at http://biodev.extra.cea.fr/strip/.
Figures
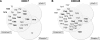



Similar articles
-
A novel non-canonical forkhead-associated (FHA) domain-binding interface mediates the interaction between Rad53 and Dbf4 proteins.J Biol Chem. 2014 Jan 31;289(5):2589-99. doi: 10.1074/jbc.M113.517060. Epub 2013 Nov 27. J Biol Chem. 2014. PMID: 24285546 Free PMC article.
-
Novel role for checkpoint Rad53 protein kinase in the initiation of chromosomal DNA replication in Saccharomyces cerevisiae.Genetics. 2006 Sep;174(1):87-99. doi: 10.1534/genetics.106.060236. Epub 2006 Jul 2. Genetics. 2006. PMID: 16816422 Free PMC article.
-
Rad53 kinase activation-independent replication checkpoint function of the N-terminal forkhead-associated (FHA1) domain.J Biol Chem. 2004 Sep 17;279(38):39636-44. doi: 10.1074/jbc.M405080200. Epub 2004 Jul 22. J Biol Chem. 2004. PMID: 15271990
-
The S-phase checkpoint: targeting the replication fork.Biol Cell. 2009 Aug 19;101(11):617-27. doi: 10.1042/BC20090053. Biol Cell. 2009. PMID: 19686094 Review.
-
Cdc7 protein kinase for DNA metabolism comes of age.Mol Microbiol. 1994 Mar;11(5):805-10. doi: 10.1111/j.1365-2958.1994.tb00358.x. Mol Microbiol. 1994. PMID: 8022258 Review.
Cited by
-
'AND' logic gates at work: Crystal structure of Rad53 bound to Dbf4 and Cdc7.Sci Rep. 2016 Sep 29;6:34237. doi: 10.1038/srep34237. Sci Rep. 2016. PMID: 27681475 Free PMC article.
-
Helicase Subunit Cdc45 Targets the Checkpoint Kinase Rad53 to Both Replication Initiation and Elongation Complexes after Fork Stalling.Mol Cell. 2019 Feb 7;73(3):562-573.e3. doi: 10.1016/j.molcel.2018.11.025. Epub 2018 Dec 27. Mol Cell. 2019. PMID: 30595439 Free PMC article.
-
Use of quantitative mass spectrometric analysis to elucidate the mechanisms of phospho-priming and auto-activation of the checkpoint kinase Rad53 in vivo.Mol Cell Proteomics. 2014 Feb;13(2):551-65. doi: 10.1074/mcp.M113.034058. Epub 2013 Dec 3. Mol Cell Proteomics. 2014. PMID: 24302356 Free PMC article.
-
Surprising complexity of the Asf1 histone chaperone-Rad53 kinase interaction.Proc Natl Acad Sci U S A. 2012 Feb 21;109(8):2866-71. doi: 10.1073/pnas.1106023109. Epub 2012 Feb 9. Proc Natl Acad Sci U S A. 2012. PMID: 22323608 Free PMC article.
-
Development of FRET biosensors for mammalian and plant systems.Protoplasma. 2014 Mar;251(2):333-47. doi: 10.1007/s00709-013-0590-z. Epub 2013 Dec 12. Protoplasma. 2014. PMID: 24337770 Review.
References
-
- Seet B. T., Dikic I., Zhou M. M., Pawson T. (2006) Reading protein modifications with interaction domains. Nat. Rev. Mol. Cell Biol. 7, 473–483 - PubMed
-
- Bhattacharyya R. P., Remenyi A., Yeh B. J., Lim W. A. (2006) Domains, Motifs, and Scaffolds: The Role of Modular Interactions in the Evolution and Wiring of Cell Signaling Circuits. Annu. Rev. Biochem. - PubMed
-
- Pawson T., Nash P. (2003) Assembly of cell regulatory systems through protein interaction domains. Science 300, 445–452 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

