The fate of metaphase kinetochores is weighed in the balance of SUMOylation during S phase
- PMID: 20724819
- PMCID: PMC3041161
- DOI: 10.4161/cc.9.16.12619
The fate of metaphase kinetochores is weighed in the balance of SUMOylation during S phase
Abstract
Genetic evidence suggests that conjugation of Small Ubiquitin-like Modifier proteins (SUMOs) plays an important role in kinetochore function, although the mechanism underlying these observations are poorly defined. We found that depletion of the SUMO protease SENP6 from HeLa cells causes chromosome misalignment, prolonged mitotic arrest and chromosome missegregation. Many inner kinetochore proteins (IKPs) were mis-localized in SENP6-depleted cells. This gross mislocalization of IKPs is due to proteolytic degradation of CENP-I and CENP-H via the SUMO targeted Ubiquitin Ligase (STUbL) pathway. Our findings show that SENP6 is a key regulator of inner kinetochore assembly that antagonizes the cellular STUbL pathway to protect IKPs from degradation during S phase. Here, we will briefly review the implications of our findings and present new data on how SUMOylation during S phase can control chromosome alignment in the subsequent metaphase.
Figures
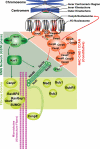

Similar articles
-
The SUMO protease SENP6 is essential for inner kinetochore assembly.J Cell Biol. 2010 Mar 8;188(5):681-92. doi: 10.1083/jcb.200909008. J Cell Biol. 2010. PMID: 20212317 Free PMC article.
-
Poly-SUMO-2/3 chain modification of Nuf2 facilitates CENP-E kinetochore localization and chromosome congression during mitosis.Cell Cycle. 2021 May;20(9):855-873. doi: 10.1080/15384101.2021.1907509. Epub 2021 Apr 28. Cell Cycle. 2021. PMID: 33910471 Free PMC article.
-
SUMO-2/3 modification and binding regulate the association of CENP-E with kinetochores and progression through mitosis.Mol Cell. 2008 Mar 28;29(6):729-41. doi: 10.1016/j.molcel.2008.01.013. Mol Cell. 2008. PMID: 18374647 Free PMC article.
-
SUMOylation in control of accurate chromosome segregation during mitosis.Curr Protein Pept Sci. 2012 Aug;13(5):467-81. doi: 10.2174/138920312802430563. Curr Protein Pept Sci. 2012. PMID: 22812528 Free PMC article. Review.
-
The SUMO Pathway in Mitosis.Adv Exp Med Biol. 2017;963:171-184. doi: 10.1007/978-3-319-50044-7_10. Adv Exp Med Biol. 2017. PMID: 28197912 Free PMC article. Review.
Cited by
-
SUMO-specific proteases/isopeptidases: SENPs and beyond.Genome Biol. 2014 Jul 31;15(7):422. doi: 10.1186/s13059-014-0422-2. Genome Biol. 2014. PMID: 25315341 Free PMC article. Review.
-
MLL5 maintains spindle bipolarity by preventing aberrant cytosolic aggregation of PLK1.J Cell Biol. 2016 Mar 28;212(7):829-43. doi: 10.1083/jcb.201501021. Epub 2016 Mar 21. J Cell Biol. 2016. PMID: 27002166 Free PMC article.
-
CSN5/JAB1 interacts with the centromeric components CENP-T and CENP-W and regulates their proteasome-mediated degradation.J Biol Chem. 2013 Sep 20;288(38):27208-27219. doi: 10.1074/jbc.M113.469221. Epub 2013 Aug 7. J Biol Chem. 2013. PMID: 23926101 Free PMC article.
-
How Does SUMO Participate in Spindle Organization?Cells. 2019 Jul 31;8(8):801. doi: 10.3390/cells8080801. Cells. 2019. PMID: 31370271 Free PMC article. Review.
-
The Smc5-Smc6 complex regulates recombination at centromeric regions and affects kinetochore protein sumoylation during normal growth.PLoS One. 2012;7(12):e51540. doi: 10.1371/journal.pone.0051540. Epub 2012 Dec 20. PLoS One. 2012. PMID: 23284708 Free PMC article.
References
-
- Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. - PubMed
-
- Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. - PubMed
-
- Hay RT. SUMO-specific proteases: a twist in the tail. Trends Cell Biol. 2007;17:370–376. - PubMed
-
- Mukhopadhyay D, Dasso M. Modification in reverse: the SUMO proteases. Trends Biochem Sci. 2007;32:286–295. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
