In vitro analysis of the Staphylococcus aureus lipoteichoic acid synthase enzyme using fluorescently labeled lipids
- PMID: 20709894
- PMCID: PMC2950504
- DOI: 10.1128/JB.00453-10
In vitro analysis of the Staphylococcus aureus lipoteichoic acid synthase enzyme using fluorescently labeled lipids
Abstract
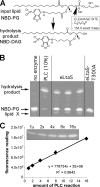
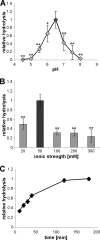
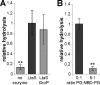
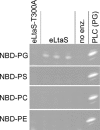
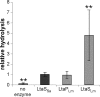
Lipoteichoic acid (LTA) is an important cell wall component of Gram-positive bacteria. The key enzyme responsible for polyglycerolphosphate lipoteichoic acid synthesis in the Gram-positive pathogen Staphylococcus aureus is the membrane-embedded lipoteichoic acid synthase enzyme, LtaS. It is presumed that LtaS hydrolyzes the glycerolphosphate head group of the membrane lipid phosphatidylglycerol (PG) and catalyzes the formation of the polyglycerolphosphate LTA backbone chain. Here we describe an in vitro assay for this new class of enzyme using PG with a fluorescently labeled fatty acid chain (NBD-PG) as the substrate and the recombinant soluble C-terminal enzymatic domain of LtaS (eLtaS). Thin-layer chromatography and mass spectrometry analysis of the lipid reaction products revealed that eLtaS is sufficient to cleave the glycerolphosphate head group from NBD-PG, resulting in the formation of NBD-diacylglycerol. An excess of soluble glycerolphosphate could not compete with the hydrolysis of the fluorescently labeled PG lipid substrate, in contrast to the addition of unlabeled PG. This indicates that the enzyme recognizes and binds other parts of the lipid substrate, besides the glycerolphosphate head group. Furthermore, eLtaS activity was Mn(2+) ion dependent; Mg(2+) and Ca(2+) supported only weak enzyme activity. Addition of Zn(2+) or EDTA inhibited enzyme activity even in the presence of Mn(2+). The pH optimum of the enzyme was 6.5, characteristic for an enzyme that functions extracellularly. Lastly, we show that the in vitro assay can be used to study the enzyme activities of other members of the lipoteichoic acid synthase enzyme family.
Figures

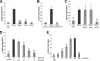
Similar articles
-
Enzymatic activities and functional interdependencies of Bacillus subtilis lipoteichoic acid synthesis enzymes.Mol Microbiol. 2011 Feb;79(3):566-83. doi: 10.1111/j.1365-2958.2010.07472.x. Epub 2010 Dec 7. Mol Microbiol. 2011. PMID: 21255105 Free PMC article.
-
Proteolytic cleavage inactivates the Staphylococcus aureus lipoteichoic acid synthase.J Bacteriol. 2011 Oct;193(19):5279-91. doi: 10.1128/JB.00369-11. Epub 2011 Jul 22. J Bacteriol. 2011. PMID: 21784926 Free PMC article.
-
Structure-based mechanism of lipoteichoic acid synthesis by Staphylococcus aureus LtaS.Proc Natl Acad Sci U S A. 2009 Feb 3;106(5):1584-9. doi: 10.1073/pnas.0809020106. Epub 2009 Jan 23. Proc Natl Acad Sci U S A. 2009. PMID: 19168632 Free PMC article.
-
Location, synthesis and function of glycolipids and polyglycerolphosphate lipoteichoic acid in Gram-positive bacteria of the phylum Firmicutes.FEMS Microbiol Lett. 2011 Jun;319(2):97-105. doi: 10.1111/j.1574-6968.2011.02260.x. Epub 2011 Mar 25. FEMS Microbiol Lett. 2011. PMID: 21388439 Free PMC article. Review.
-
Lipoteichoic acid synthesis and function in gram-positive bacteria.Annu Rev Microbiol. 2014;68:81-100. doi: 10.1146/annurev-micro-091213-112949. Epub 2014 May 5. Annu Rev Microbiol. 2014. PMID: 24819367 Review.
Cited by
-
The essential inner membrane protein YejM is a metalloenzyme.Sci Rep. 2020 Oct 20;10(1):17794. doi: 10.1038/s41598-020-73660-6. Sci Rep. 2020. PMID: 33082366 Free PMC article.
-
Membrane proteases in the bacterial protein secretion and quality control pathway.Microbiol Mol Biol Rev. 2012 Jun;76(2):311-30. doi: 10.1128/MMBR.05019-11. Microbiol Mol Biol Rev. 2012. PMID: 22688815 Free PMC article. Review.
-
Staphylococcus aureus Extracellular Vesicles Elicit an Immunostimulatory Response in vivo on the Murine Mammary Gland.Front Cell Infect Microbiol. 2018 Aug 22;8:277. doi: 10.3389/fcimb.2018.00277. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 30186772 Free PMC article.
-
Enzymatic activities and functional interdependencies of Bacillus subtilis lipoteichoic acid synthesis enzymes.Mol Microbiol. 2011 Feb;79(3):566-83. doi: 10.1111/j.1365-2958.2010.07472.x. Epub 2010 Dec 7. Mol Microbiol. 2011. PMID: 21255105 Free PMC article.
-
A Novel Antimicrobial Mechanism of Azalomycin F Acting on Lipoteichoic Acid Synthase and Cell Envelope.Molecules. 2024 Feb 14;29(4):856. doi: 10.3390/molecules29040856. Molecules. 2024. PMID: 38398608 Free PMC article.
References
-
- Abachin, E., C. Poyart, E. Pellegrini, E. Milohanic, F. Fiedler, P. Berche, and P. Trieu-Cuot. 2002. Formation of d-alanyl-lipoteichoic acid is required for adhesion and virulence of Listeria monocytogenes. Mol. Microbiol. 43:1-14. - PubMed
-
- Archibald, A. R., J. J. Armstrong, J. Baddiley, and J. B. Hay. 1961. Teichoic acids and the structure of bacterial walls. Nature 191:570-572. - PubMed
-
- Collins, L. V., S. A. Kristian, C. Weidenmaier, M. Faigle, K. P. Van Kessel, J. A. Van Strijp, F. Götz, B. Neumeister, and A. Peschel. 2002. Staphylococcus aureus strains lacking d-alanine modifications of teichoic acids are highly susceptible to human neutrophil killing and are virulence attenuated in mice. J. Infect. Dis. 186:214-219. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

