Downstream of identity genes: muscle-type-specific regulation of the fusion process
- PMID: 20708593
- PMCID: PMC3852356
- DOI: 10.1016/j.devcel.2010.07.008
Downstream of identity genes: muscle-type-specific regulation of the fusion process
Abstract
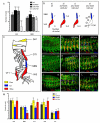
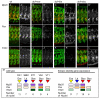
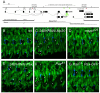
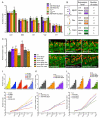
In all metazoan organisms, the diversification of cell types involves determination of cell fates and subsequent execution of specific differentiation programs. During Drosophila myogenesis, identity genes specify the fates of founder myoblasts, from which derive all individual larval muscles. Here, to understand how cell fate information residing within founders is translated during differentiation, we focus on three identity genes, eve, lb, and slou, and how they control the size of individual muscles by regulating the number of fusion events. They achieve this by setting expression levels of Mp20, Pax, and mspo, three genes that regulate actin dynamics and cell adhesion and, as we show here, modulate the fusion process in a muscle-specific manner. Thus, these data show how the identity information implemented by transcription factors is translated via target genes into cell-type-specific programs of differentiation.
2010 Elsevier Inc. All rights reserved.
Figures



Similar articles
-
Founder myoblasts and fibre number during adult myogenesis in Drosophila.Development. 2004 Aug;131(15):3761-72. doi: 10.1242/dev.01249. Development. 2004. PMID: 15262890
-
Org-1, the Drosophila ortholog of Tbx1, is a direct activator of known identity genes during muscle specification.Development. 2012 Mar;139(5):1001-12. doi: 10.1242/dev.073890. Development. 2012. PMID: 22318630 Free PMC article.
-
Mind bomb 2, a founder myoblast-specific protein, regulates myoblast fusion and muscle stability.Development. 2008 Mar;135(5):849-57. doi: 10.1242/dev.015529. Epub 2008 Jan 23. Development. 2008. PMID: 18216171
-
Visualizing new dimensions in Drosophila myoblast fusion.Bioessays. 2008 May;30(5):423-31. doi: 10.1002/bies.20756. Bioessays. 2008. PMID: 18404690 Free PMC article. Review.
-
Diversification of muscle types in Drosophila embryos.Exp Cell Res. 2022 Jan 1;410(1):112950. doi: 10.1016/j.yexcr.2021.112950. Epub 2021 Nov 26. Exp Cell Res. 2022. PMID: 34838813 Review.
Cited by
-
Specification of the somatic musculature in Drosophila.Wiley Interdiscip Rev Dev Biol. 2015 Jul-Aug;4(4):357-75. doi: 10.1002/wdev.182. Epub 2015 Feb 27. Wiley Interdiscip Rev Dev Biol. 2015. PMID: 25728002 Free PMC article. Review.
-
Distinct roles for Ste20-like kinase SLK in muscle function and regeneration.Skelet Muscle. 2013 Jul 1;3(1):16. doi: 10.1186/2044-5040-3-16. Skelet Muscle. 2013. PMID: 23815977 Free PMC article.
-
Novel functions for integrin-associated proteins revealed by analysis of myofibril attachment in Drosophila.Elife. 2018 Jul 20;7:e35783. doi: 10.7554/eLife.35783. Elife. 2018. PMID: 30028294 Free PMC article.
-
Muscle cell fate choice requires the T-box transcription factor midline in Drosophila.Genetics. 2015 Mar;199(3):777-91. doi: 10.1534/genetics.115.174300. Epub 2015 Jan 21. Genetics. 2015. PMID: 25614583 Free PMC article.
-
The genetic architecture of temperature adaptation is shaped by population ancestry and not by selection regime.Genome Biol. 2021 Jul 16;22(1):211. doi: 10.1186/s13059-021-02425-9. Genome Biol. 2021. PMID: 34271951 Free PMC article.
References
-
- Abmayr SM, Zhuang S, Geisbrecht ER. Myoblast fusion in Drosophila. Methods Mol Biol. 2008;475:75–97. - PubMed
-
- Artero R, Furlong EE, Beckett K, Scott MP, Baylies M. Notch and Ras signaling pathway effector genes expressed in fusion competent and founder cells during Drosophila myogenesis. Development. 2003;130:6257–72. - PubMed
-
- Bate M. The embryonic development of larval muscles in Drosophila. Development. 1990;110:791–804. - PubMed
-
- Baylies MK, Michelson AM. Invertebrate myogenesis: looking back to the future of muscle development. Curr Opin Genet Dev. 2001;11:431–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

