Cdk phosphorylation of a nucleoporin controls localization of active genes through the cell cycle
- PMID: 20702586
- PMCID: PMC2947477
- DOI: 10.1091/mbc.E10-01-0065
Cdk phosphorylation of a nucleoporin controls localization of active genes through the cell cycle
Abstract
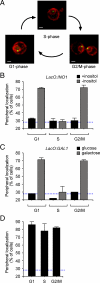
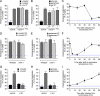
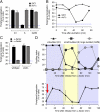
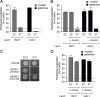
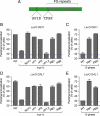
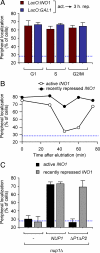
Many inducible genes in yeast are targeted to the nuclear pore complex when active. We find that the peripheral localization of the INO1 and GAL1 genes is regulated through the cell cycle. Active INO1 and GAL1 localized at the nuclear periphery during G1, became nucleoplasmic during S-phase, and then returned to the nuclear periphery during G2/M. Loss of peripheral targeting followed the initiation of DNA replication and was lost in cells lacking a cyclin-dependent kinase (Cdk) inhibitor. Furthermore, the Cdk1 kinase and two Cdk phosphorylation sites in the nucleoporin Nup1 were required for peripheral targeting of INO1 and GAL1. Introduction of aspartic acid residues in place of either of these two sites in Nup1 bypassed the requirement for Cdk1 and resulted in targeting of INO1 and GAL1 to the nuclear periphery during S-phase. Thus, phosphorylation of a nuclear pore component by cyclin dependent kinase controls the localization of active genes to the nuclear periphery through the cell cycle.
Figures
Similar articles
-
Gene positioning is regulated by phosphorylation of the nuclear pore complex by Cdk1.Cell Cycle. 2011 Feb 1;10(3):392-5. doi: 10.4161/cc.10.3.14644. Epub 2011 Feb 1. Cell Cycle. 2011. PMID: 21228627 Free PMC article.
-
Interaction of a DNA zip code with the nuclear pore complex promotes H2A.Z incorporation and INO1 transcriptional memory.Mol Cell. 2010 Oct 8;40(1):112-25. doi: 10.1016/j.molcel.2010.09.007. Mol Cell. 2010. PMID: 20932479 Free PMC article.
-
Epigenetic Transcriptional Memory of GAL Genes Depends on Growth in Glucose and the Tup1 Transcription Factor in Saccharomyces cerevisiae.Genetics. 2017 Aug;206(4):1895-1907. doi: 10.1534/genetics.117.201632. Epub 2017 Jun 12. Genetics. 2017. PMID: 28607146 Free PMC article.
-
Transcriptional memory at the nuclear periphery.Curr Opin Cell Biol. 2009 Feb;21(1):127-33. doi: 10.1016/j.ceb.2009.01.007. Epub 2009 Jan 30. Curr Opin Cell Biol. 2009. PMID: 19181512 Free PMC article. Review.
-
[Molecular mechanisms controlling the cell cycle: fundamental aspects and implications for oncology].Cancer Radiother. 2001 Apr;5(2):109-29. doi: 10.1016/s1278-3218(01)00087-7. Cancer Radiother. 2001. PMID: 11355576 Review. French.
Cited by
-
Nuclear pore complexes and regulation of gene expression.Curr Opin Cell Biol. 2017 Jun;46:26-32. doi: 10.1016/j.ceb.2016.12.006. Epub 2017 Jan 11. Curr Opin Cell Biol. 2017. PMID: 28088069 Free PMC article. Review.
-
A change in nuclear pore complex composition regulates cell differentiation.Dev Cell. 2012 Feb 14;22(2):446-58. doi: 10.1016/j.devcel.2011.11.021. Epub 2012 Jan 19. Dev Cell. 2012. PMID: 22264802 Free PMC article.
-
Phosphorylation-dependent mitotic SUMOylation drives nuclear envelope-chromatin interactions.J Cell Biol. 2021 Dec 6;220(12):e202103036. doi: 10.1083/jcb.202103036. Epub 2021 Nov 17. J Cell Biol. 2021. PMID: 34787675 Free PMC article.
-
Principles of chromosomal organization: lessons from yeast.J Cell Biol. 2011 Mar 7;192(5):723-33. doi: 10.1083/jcb.201010058. J Cell Biol. 2011. PMID: 21383075 Free PMC article. Review.
-
Gene positioning and expression.Curr Opin Cell Biol. 2011 Jun;23(3):338-45. doi: 10.1016/j.ceb.2011.01.001. Epub 2011 Feb 1. Curr Opin Cell Biol. 2011. PMID: 21292462 Free PMC article. Review.
References
-
- Berger A. B., Cabal G. G., Fabre E., Duong T., Buc H., Nehrbass U., Olivo-Marin J. C., Gadal O., Zimmer C. High-resolution statistical mapping reveals gene territories in live yeast. Nat. Methods. 2008;5:1031–1037. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

