Coordination of substrate binding and ATP hydrolysis in Vps4-mediated ESCRT-III disassembly
- PMID: 20702581
- PMCID: PMC2947475
- DOI: 10.1091/mbc.E10-06-0512
Coordination of substrate binding and ATP hydrolysis in Vps4-mediated ESCRT-III disassembly
Abstract
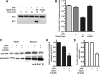
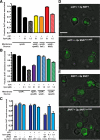
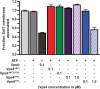
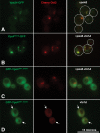
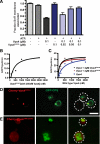
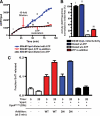
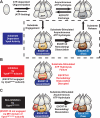
ESCRT-III undergoes dynamic assembly and disassembly to facilitate membrane exvagination processes including multivesicular body (MVB) formation, enveloped virus budding, and membrane abscission during cytokinesis. The AAA-ATPase Vps4 is required for ESCRT-III disassembly, however the coordination of Vps4 ATP hydrolysis with ESCRT-III binding and disassembly is not understood. Vps4 ATP hydrolysis has been proposed to execute ESCRT-III disassembly as either a stable oligomer or an unstable oligomer whose dissociation drives ESCRT-III disassembly. An in vitro ESCRT-III disassembly assay was developed to analyze Vps4 function during this process. The studies presented here support a model in which Vps4 acts as a stable oligomer during ATP hydrolysis and ESCRT-III disassembly. Moreover, Vps4 oligomer binding to ESCRT-III induces coordination of ATP hydrolysis at the level of individual Vps4 subunits. These results suggest that Vps4 functions as a stable oligomer that acts upon individual ESCRT-III subunits to facilitate ESCRT-III disassembly.
Figures

Similar articles
-
Binding of Substrates to the Central Pore of the Vps4 ATPase Is Autoinhibited by the Microtubule Interacting and Trafficking (MIT) Domain and Activated by MIT Interacting Motifs (MIMs).J Biol Chem. 2015 May 22;290(21):13490-9. doi: 10.1074/jbc.M115.642355. Epub 2015 Apr 1. J Biol Chem. 2015. PMID: 25833946 Free PMC article.
-
Vps4 stimulatory element of the cofactor Vta1 contacts the ATPase Vps4 α7 and α9 to stimulate ATP hydrolysis.J Biol Chem. 2014 Oct 10;289(41):28707-18. doi: 10.1074/jbc.M114.580696. Epub 2014 Aug 27. J Biol Chem. 2014. PMID: 25164817 Free PMC article.
-
Conformational Changes in the Endosomal Sorting Complex Required for the Transport III Subunit Ist1 Lead to Distinct Modes of ATPase Vps4 Regulation.J Biol Chem. 2015 Dec 11;290(50):30053-65. doi: 10.1074/jbc.M115.665604. Epub 2015 Oct 29. J Biol Chem. 2015. PMID: 26515066 Free PMC article.
-
The role of VPS4 in ESCRT-III polymer remodeling.Biochem Soc Trans. 2019 Feb 28;47(1):441-448. doi: 10.1042/BST20180026. Epub 2019 Feb 19. Biochem Soc Trans. 2019. PMID: 30783012 Review.
-
Structures, Functions, and Dynamics of ESCRT-III/Vps4 Membrane Remodeling and Fission Complexes.Annu Rev Cell Dev Biol. 2018 Oct 6;34:85-109. doi: 10.1146/annurev-cellbio-100616-060600. Epub 2018 Aug 10. Annu Rev Cell Dev Biol. 2018. PMID: 30095293 Free PMC article. Review.
Cited by
-
An ESCRT/VPS4 Envelopment Trap To Examine the Mechanism of Alphaherpesvirus Assembly and Transport in Neurons.J Virol. 2022 Mar 23;96(6):e0217821. doi: 10.1128/jvi.02178-21. Epub 2022 Jan 19. J Virol. 2022. PMID: 35045266 Free PMC article.
-
HIV-1 assembly, budding, and maturation.Cold Spring Harb Perspect Med. 2012 Jul;2(7):a006924. doi: 10.1101/cshperspect.a006924. Cold Spring Harb Perspect Med. 2012. PMID: 22762019 Free PMC article. Review.
-
Insights into the function of ESCRT complex and LBPA in ASFV infection.Front Cell Infect Microbiol. 2023 Dec 6;13:1163569. doi: 10.3389/fcimb.2023.1163569. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 38125905 Free PMC article.
-
Inhibition of HBV replication by VPS4B and its dominant negative mutant VPS4B-K180Q in vivo.J Huazhong Univ Sci Technolog Med Sci. 2012 Jun;32(3):311-316. doi: 10.1007/s11596-012-0054-2. Epub 2012 Jun 9. J Huazhong Univ Sci Technolog Med Sci. 2012. PMID: 22684550
-
Endosomal Na+ (K+)/H+ exchanger Nhx1/Vps44 functions independently and downstream of multivesicular body formation.J Biol Chem. 2011 Dec 23;286(51):44067-44077. doi: 10.1074/jbc.M111.282319. Epub 2011 Oct 13. J Biol Chem. 2011. PMID: 21998311 Free PMC article.
References
-
- Azmi I. F., Davies B. A., Xiao J., Babst M., Xu Z., Katzmann D. J. ESCRT-III family members stimulate Vps4 ATPase activity directly or via Vta1. Dev. Cell. 2008;14:50–61. - PubMed
-
- Babst M., Katzmann D. J., Estepa-Sabal E. J., Meerloo T., Emr S. D. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell. 2002a;3:271–282. - PubMed
-
- Babst M., Katzmann D. J., Snyder W. B., Wendland B., Emr S. D. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell. 2002b;3:283–289. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

