Trisomic dose of several chromosome 21 genes perturbs haematopoietic stem and progenitor cell differentiation in Down's syndrome
- PMID: 20697343
- PMCID: PMC3007620
- DOI: 10.1038/onc.2010.351
Trisomic dose of several chromosome 21 genes perturbs haematopoietic stem and progenitor cell differentiation in Down's syndrome
Abstract
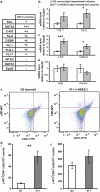
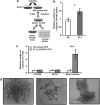
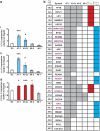
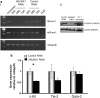
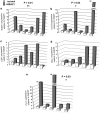
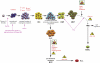
Children with Down's syndrome (DS) have 20-50-fold higher incidence of all leukaemias (lymphoid and myeloid), for reasons not understood. As incidence of many solid tumours is much lower in DS, we speculated that disturbed early haematopoietic differentiation could be the cause of increased leukaemia risk. If a common mechanism is behind the risk of both major leukaemia types, it would have to arise before the bifurcation to myeloid and lymphoid lineages. Using the transchromosomic system (mouse embryonic stem cells (ESCs)) bearing an extra human chromosome 21 (HSA21)) we analyzed the early stages of haematopoietic commitment (mesodermal colony formation) in vitro. We observed that trisomy 21 (T21) causes increased production of haemogenic endothelial cells, haematopoietic stem cell precursors and increased colony forming potential, with significantly increased immature progenitors. Transchromosomic colonies showed increased expression of Gata-2, c-Kit and Tie-2. A panel of partial T21 ESCs allowed us to assign these effects to HSA21 sub-regions, mapped by 3.5 kbp-resolution tiling arrays. The Gata-2 increase on one side, and c-Kit and Tie-2 increases on the other, could be attributed to two different, non-overlapping HSA21 regions. Using human-specific small interfering RNA silencing, we could demonstrate that an extra copy of RUNX1, but not ETS-2 or ERG, causes an increase in Tie-2/c-Kit levels. Finally, we detected significantly increased levels of RUNX1, C-KIT and PU.1 in human foetal livers with T21. We conclude that overdose of more than one HSA21 gene contributes to the disturbance of early haematopoiesis in DS, and that one of the contributors is RUNX1. As the observed T21-driven hyperproduction of multipotential immature precursors precedes the bifurcation to lymphoid and myeloid lineages, we speculate that this could create conditions of increased chance for acquisition of pre-leukaemogenic rearrangements/mutations in both lymphoid and myeloid lineages during foetal haematopoiesis, contributing to the increased risk of both leukaemia types in DS.
Figures
Comment in
-
Trisomy 21 leukemias: finding the hits that matter.Oncogene. 2010 Nov 18;29(46):6099-101. doi: 10.1038/onc.2010.353. Epub 2010 Aug 16. Oncogene. 2010. PMID: 20711238
Similar articles
-
The impact of trisomy 21 on foetal haematopoiesis.Blood Cells Mol Dis. 2013 Dec;51(4):277-81. doi: 10.1016/j.bcmd.2013.07.008. Epub 2013 Aug 7. Blood Cells Mol Dis. 2013. PMID: 23932236 Free PMC article. Review.
-
[The investigation of hematopoietic capacity of HPP-CFC derived from murine embryonic stem cells in vitro and in vivo].Sheng Wu Gong Cheng Xue Bao. 2003 May;19(3):312-6. Sheng Wu Gong Cheng Xue Bao. 2003. PMID: 15969013 Chinese.
-
Early lineage priming by trisomy of Erg leads to myeloproliferation in a Down syndrome model.PLoS Genet. 2015 May 14;11(5):e1005211. doi: 10.1371/journal.pgen.1005211. eCollection 2015 May. PLoS Genet. 2015. PMID: 25973911 Free PMC article.
-
Highly penetrant myeloproliferative disease in the Ts65Dn mouse model of Down syndrome.Blood. 2008 Jan 15;111(2):767-75. doi: 10.1182/blood-2007-04-085670. Epub 2007 Sep 27. Blood. 2008. PMID: 17901249 Free PMC article.
-
Haematopoietic development and leukaemia in Down syndrome.Br J Haematol. 2014 Dec;167(5):587-99. doi: 10.1111/bjh.13096. Epub 2014 Aug 22. Br J Haematol. 2014. PMID: 25155832 Review.
Cited by
-
Modeling Down Syndrome Myeloid Leukemia by Sequential Introduction of GATA1 and STAG2 Mutations in Induced Pluripotent Stem Cells with Trisomy 21.Cells. 2022 Feb 11;11(4):628. doi: 10.3390/cells11040628. Cells. 2022. PMID: 35203280 Free PMC article.
-
Down syndrome iPSC model: endothelial perspective on tumor development.Oncotarget. 2020 Sep 8;11(36):3387-3404. doi: 10.18632/oncotarget.27712. eCollection 2020 Sep 8. Oncotarget. 2020. PMID: 32934781 Free PMC article.
-
Abnormal microRNA expression in Ts65Dn hippocampus and whole blood: contributions to Down syndrome phenotypes.Dev Neurosci. 2011;33(5):451-67. doi: 10.1159/000330884. Epub 2011 Oct 27. Dev Neurosci. 2011. PMID: 22042248 Free PMC article.
-
Impaired Iron Homeostasis and Haematopoiesis Impacts Inflammation in the Ageing Process in Down Syndrome Dementia.J Clin Med. 2021 Jun 29;10(13):2909. doi: 10.3390/jcm10132909. J Clin Med. 2021. PMID: 34209847 Free PMC article.
-
Down syndrome and leukemia: insights into leukemogenesis and translational targets.Transl Pediatr. 2015 Apr;4(2):76-92. doi: 10.3978/j.issn.2224-4336.2015.03.03. Transl Pediatr. 2015. PMID: 26835364 Free PMC article. Review.
References
-
- Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. - PubMed
-
- Al-Kasim F, Doyle JJ, Massey GV, Weinstein HJ, Zipursky A. Incidence and treatment of potentially lethal diseases in transient leukemia of Down syndrome: Pediatric Oncology Group Study. J Pediatr Hematol Oncol. 2002;24:9–13. - PubMed
-
- Bahn S, Mimmack M, Ryan M, Caldwell MA, Jauniaux E, Starkey M, et al. Neuronal target genes of the neuron-restrictive silencer factor in neurospheres derived from fetuses with Down's syndrome: a gene expression study. Lancet. 2002;359:310–315. - PubMed
-
- Bercovich D, Ganmore I, Scott LM, Wainreb G, Birger Y, Elimelech A, et al. Mutations of JAK2 in acute lymphoblastic leukaemias associated with Down's syndrome. Lancet. 2008;372:1484–1492. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

