Activation and repression of glucose-stimulated ChREBP requires the concerted action of multiple domains within the MondoA conserved region
- PMID: 20682844
- PMCID: PMC2957870
- DOI: 10.1152/ajpendo.00349.2010
Activation and repression of glucose-stimulated ChREBP requires the concerted action of multiple domains within the MondoA conserved region
Abstract
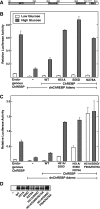
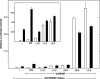
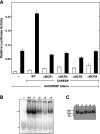
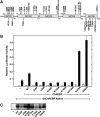
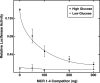
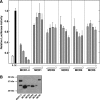
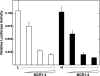
Carbohydrate response element-binding protein (ChREBP) is a glucose-dependent transcription factor that stimulates the expression of glycolytic and lipogenic genes in mammals. Glucose regulation of ChREBP has been mapped to its conserved NH(2)-terminal region of 300 amino acids, designated the MondoA conserved region (MCR). Within the MCR, five domains (MCR1-5) have a particularly high level of conservation and are likely to be important for glucose regulation. We carried out a large-scale deletion and substitution mutational analysis of the MCR domain of ChREBP. This analysis revealed that MCRs 1-4 function in a concerted fashion to repress ChREBP activity in basal (nonstimulatory) conditions. Deletion of the entire MCR1-4 segment or the combination of four specific point mutations located across this region leads to a highly active, glucose-independent form of ChREBP. However, deletion of any individual MCR domain and the majority of point mutations throughout MCR1-4 rendered ChREBP inactive. These observations suggest that the MCR1-4 region interacts with an additional coregulatory factor required for activation. This possibility is supported by the observation that the MCR1-4 region can compete for activity with wild-type ChREBP in stimulatory conditions. In contrast, mutations in the MCR5 domain result in increased activity, suggesting that this domain may be the target of intramolecular repression in basal conditions. Thus, the MCR domains act in a complex and coordinated manner to regulate ChREBP activity in response to glucose.
Figures

Similar articles
-
Glucose activates ChREBP by increasing its rate of nuclear entry and relieving repression of its transcriptional activity.J Biol Chem. 2008 Aug 29;283(35):24029-38. doi: 10.1074/jbc.M801539200. Epub 2008 Jun 30. J Biol Chem. 2008. PMID: 18591247 Free PMC article.
-
Glucose-mediated transactivation of carbohydrate response element-binding protein requires cooperative actions from Mondo conserved regions and essential trans-acting factor 14-3-3.Mol Endocrinol. 2008 Jul;22(7):1658-72. doi: 10.1210/me.2007-0560. Epub 2008 Apr 24. Mol Endocrinol. 2008. PMID: 18436566 Free PMC article.
-
A novel N-terminal domain may dictate the glucose response of Mondo proteins.PLoS One. 2012;7(4):e34803. doi: 10.1371/journal.pone.0034803. Epub 2012 Apr 10. PLoS One. 2012. PMID: 22506051 Free PMC article.
-
Glucose-Sensing Transcription Factor MondoA/ChREBP as Targets for Type 2 Diabetes: Opportunities and Challenges.Int J Mol Sci. 2019 Oct 16;20(20):5132. doi: 10.3390/ijms20205132. Int J Mol Sci. 2019. PMID: 31623194 Free PMC article. Review.
-
Glucose sensing by ChREBP/MondoA-Mlx transcription factors.Semin Cell Dev Biol. 2012 Aug;23(6):640-7. doi: 10.1016/j.semcdb.2012.02.007. Epub 2012 Mar 3. Semin Cell Dev Biol. 2012. PMID: 22406740 Review.
Cited by
-
Normal and Neoplastic Growth Suppression by the Extended Myc Network.Cells. 2022 Feb 21;11(4):747. doi: 10.3390/cells11040747. Cells. 2022. PMID: 35203395 Free PMC article. Review.
-
Functional interactions among members of the MAX and MLX transcriptional network during oncogenesis.Biochim Biophys Acta. 2015 May;1849(5):484-500. doi: 10.1016/j.bbagrm.2014.05.016. Epub 2014 May 22. Biochim Biophys Acta. 2015. PMID: 24857747 Free PMC article. Review.
-
Induction of the ChREBPβ Isoform Is Essential for Glucose-Stimulated β-Cell Proliferation.Diabetes. 2015 Dec;64(12):4158-70. doi: 10.2337/db15-0239. Epub 2015 Sep 17. Diabetes. 2015. PMID: 26384380 Free PMC article.
-
The polyol pathway is an evolutionarily conserved system for sensing glucose uptake.PLoS Biol. 2022 Jun 10;20(6):e3001678. doi: 10.1371/journal.pbio.3001678. eCollection 2022 Jun. PLoS Biol. 2022. PMID: 35687590 Free PMC article.
-
Transcriptional control of hepatic lipid metabolism by SREBP and ChREBP.Semin Liver Dis. 2013 Nov;33(4):301-11. doi: 10.1055/s-0033-1358523. Epub 2013 Nov 12. Semin Liver Dis. 2013. PMID: 24222088 Free PMC article. Review.
References
-
- Billin AN, Eilers AL, Queva C, Ayer DE. Mlx, a novel Max-like BHLHZip protein that interacts with the Max network of transcription factors. J Biol Chem 274: 36344–36350, 1999 - PubMed
-
- Collier JJ, Zhang P, Pedersen KB, Burke SJ, Haycock JW, Scott DK. c-Myc and ChREBP regulate glucose-mediated expression of the L-type pyruvate kinase gene in INS-1-derived 832/13 cells. Am J Physiol Endocrinol Metab 293: E48–E56, 2007 - PubMed
-
- da Silva Xavier G, Rutter GA, Diraison F, Andreolas C, Leclerc I. ChREBP binding to fatty acid synthase and L-type pyruvate kinase genes is stimulated by glucose in pancreatic beta-cells. J Lipid Res 47: 2482–2491, 2006 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

