3'-coterminal subgenomic RNAs and putative cis-acting elements of Grapevine leafroll-associated virus 3 reveals 'unique' features of gene expression strategy in the genus Ampelovirus
- PMID: 20682046
- PMCID: PMC2922190
- DOI: 10.1186/1743-422X-7-180
3'-coterminal subgenomic RNAs and putative cis-acting elements of Grapevine leafroll-associated virus 3 reveals 'unique' features of gene expression strategy in the genus Ampelovirus
Abstract
Background: The family Closteroviridae comprises genera with monopartite genomes, Closterovirus and Ampelovirus, and with bipartite and tripartite genomes, Crinivirus. By contrast to closteroviruses in the genera Closterovirus and Crinivirus, much less is known about the molecular biology of viruses in the genus Ampelovirus, although they cause serious diseases in agriculturally important perennial crops like grapevines, pineapple, cherries and plums.
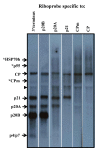
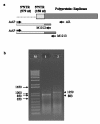
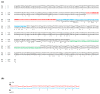
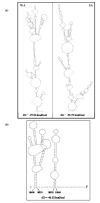
Results: The gene expression and cis-acting elements of Grapevine leafroll-associated virus 3 (GLRaV-3; genus Ampelovirus) was examined and compared to that of other members of the family Closteroviridae. Six putative 3'-coterminal subgenomic (sg) RNAs were abundantly present in grapevine (Vitis vinifera) infected with GLRaV-3. The sgRNAs for coat protein (CP), p21, p20A and p20B were confirmed using gene-specific riboprobes in Northern blot analysis. The 5'-termini of sgRNAs specific to CP, p21, p20A and p20B were mapped in the 18,498 nucleotide (nt) virus genome and their leader sequences determined to be 48, 23, 95 and 125 nt, respectively. No conserved motifs were found around the transcription start site or in the leader sequence of these sgRNAs. The predicted secondary structure analysis of sequences around the start site failed to reveal any conserved motifs among the four sgRNAs. The GLRaV-3 isolate from Washington had a 737 nt long 5' nontranslated region (NTR) with a tandem repeat of 65 nt sequence and differed in sequence and predicted secondary structure with a South Africa isolate. Comparison of the dissimilar sequences of the 5'NTRs did not reveal any common predicted structures. The 3'NTR was shorter and more conserved. The lack of similarity among the cis-acting elements of the diverse viruses in the family Closteroviridae is another measure of the complexity of their evolution.
Conclusions: The results indicate that transcription regulation of GLRaV-3 sgRNAs appears to be different from members of the genus Closterovirus. An analysis of the genome sequence confirmed that GLRaV-3 has an unusually long 5'NTR of 737 nt compared to other monopartite members of the family Closteroviridae, with distinct differences in the sequence and predicted secondary structure when compared to the corresponding region of the GLRaV-3 isolate from South Africa.
Figures


Similar articles
-
An Analysis of the Complete Genome Sequence and Subgenomic RNAs Reveals Unique Features of the Ampelovirus, Grapevine leafroll-associated virus 1.Phytopathology. 2017 Sep;107(9):1069-1079. doi: 10.1094/PHYTO-02-17-0061-R. Epub 2017 Jul 7. Phytopathology. 2017. PMID: 28686140
-
Intra-species recombination among strains of the ampelovirus Grapevine leafroll-associated virus 4.Virol J. 2019 Nov 19;16(1):139. doi: 10.1186/s12985-019-1243-4. Virol J. 2019. PMID: 31744534 Free PMC article.
-
Genomic analysis of grapevine leafroll associated virus-5 and related viruses.Virus Res. 2012 Jan;163(1):19-27. doi: 10.1016/j.virusres.2011.08.006. Epub 2011 Aug 30. Virus Res. 2012. PMID: 21893115
-
Probing into the Effects of Grapevine Leafroll-Associated Viruses on the Physiology, Fruit Quality and Gene Expression of Grapes.Viruses. 2021 Mar 31;13(4):593. doi: 10.3390/v13040593. Viruses. 2021. PMID: 33807294 Free PMC article. Review.
-
Grapevine leafroll disease and associated viruses: a unique pathosystem.Annu Rev Phytopathol. 2015;53:613-34. doi: 10.1146/annurev-phyto-102313-045946. Annu Rev Phytopathol. 2015. PMID: 26243729 Review.
Cited by
-
A divergent variant of Grapevine leafroll-associated virus 3 is present in California.Virol J. 2012 Oct 13;9:235. doi: 10.1186/1743-422X-9-235. Virol J. 2012. PMID: 23062082 Free PMC article.
-
Modulation of flavonoid biosynthetic pathway genes and anthocyanins due to virus infection in grapevine (Vitis vinifera L.) leaves.BMC Plant Biol. 2010 Aug 23;10:187. doi: 10.1186/1471-2229-10-187. BMC Plant Biol. 2010. PMID: 20731850 Free PMC article.
-
Analysis of pineapple mealybug wilt associated virus -1 and -2 for potential RNA silencing suppressors and pathogenicity factors.Viruses. 2015 Mar 5;7(3):969-95. doi: 10.3390/v7030969. Viruses. 2015. PMID: 25751306 Free PMC article.
-
Real-time RT-PCR high-resolution melting curve analysis and multiplex RT-PCR to detect and differentiate grapevine leafroll-associated virus 3 variant groups I, II, III and VI.Virol J. 2012 Sep 27;9:219. doi: 10.1186/1743-422X-9-219. Virol J. 2012. PMID: 23016734 Free PMC article.
-
Next-Generation Sequencing Combined With Conventional Sanger Sequencing Reveals High Molecular Diversity in Actinidia Virus 1 Populations From Kiwifruit Grown in China.Front Microbiol. 2020 Dec 16;11:602039. doi: 10.3389/fmicb.2020.602039. eCollection 2020. Front Microbiol. 2020. PMID: 33391218 Free PMC article.
References
-
- Salem NM, Chen AY, Tzanetakis IE, Mongkolsiriwattana C, Ng JC. Further complexity of the genus Crinivirus revealed by the complete genome sequence of Lettuce chlorosis virus (LCV) and the similar temporal accumulation of LCV genomic RNAs 1 and 2. Virology. 2009;390:45–55. doi: 10.1016/j.virol.2009.04.025. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

