SAP protein-dependent natural killer T-like cells regulate the development of CD8(+) T cells with innate lymphocyte characteristics
- PMID: 20674402
- PMCID: PMC2933745
- DOI: 10.1016/j.immuni.2010.07.013
SAP protein-dependent natural killer T-like cells regulate the development of CD8(+) T cells with innate lymphocyte characteristics
Abstract
CD8(+) T cells are selected via low-affinity interaction with MHC class I molecules on thymic epithelial cells (TECs). However, compromised T cell receptor signaling was proposed to force CD8(+) T cell selection on hematopoietic cells through a SLAM-associated protein (SAP)-dependent mechanism similar to NKT cells. The outcome is an unconventional CD8(+) T cell with phenotypic and functional characteristics of innate lymphocytes. Here we showed that Id3(-/-) CD8(+) T cells had an innate-like phenotype and required SAP for their development. However, like conventional CD8(+) T cells, Id3(-/-) CD8(+) thymocytes were selected on TECs. The requirement for SAP and the innate-like phenotype was not intrinsic to Id3(-/-) CD8(+) thymocytes. Rather, an expanded population of NKT-like cells induced the innate phenotype on CD8(+) T cells through production of interleukin-4. Our findings reveal that accumulation of NKT-like cells promotes conventional CD8(+) thymocytes to acquire innate lymphocyte characteristics.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures

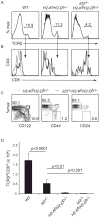
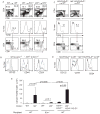
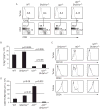



Comment in
-
NKT cells favour the unconventional.Nat Rev Immunol. 2010 Sep;10(9):618. doi: 10.1038/nri2834. Nat Rev Immunol. 2010. PMID: 21080587 No abstract available.
Similar articles
-
Development of innate CD4+ and CD8+ T cells in Itk-deficient mice is regulated by distinct pathways.J Immunol. 2014 Jul 15;193(2):688-99. doi: 10.4049/jimmunol.1302059. Epub 2014 Jun 18. J Immunol. 2014. PMID: 24943215 Free PMC article.
-
Inhibitor of DNA binding 3 limits development of murine slam-associated adaptor protein-dependent "innate" gammadelta T cells.PLoS One. 2010 Feb 19;5(2):e9303. doi: 10.1371/journal.pone.0009303. PLoS One. 2010. PMID: 20174563 Free PMC article.
-
Cutting edge: Ly9 (CD229), a SLAM family receptor, negatively regulates the development of thymic innate memory-like CD8+ T and invariant NKT cells.J Immunol. 2013 Jan 1;190(1):21-6. doi: 10.4049/jimmunol.1202435. Epub 2012 Dec 7. J Immunol. 2013. PMID: 23225888 Free PMC article.
-
The Role of Adaptor Proteins in the Biology of Natural Killer T (NKT) Cells.Front Immunol. 2019 Jun 25;10:1449. doi: 10.3389/fimmu.2019.01449. eCollection 2019. Front Immunol. 2019. PMID: 31293596 Free PMC article. Review.
-
Consequence of the SLAM-SAP signaling pathway in innate-like and conventional lymphocytes.Immunity. 2007 Nov;27(5):698-710. doi: 10.1016/j.immuni.2007.11.005. Immunity. 2007. PMID: 18031694 Review.
Cited by
-
NFAT2 Regulates Generation of Innate-Like CD8+ T Lymphocytes and CD8+ T Lymphocytes Responses.Front Immunol. 2016 Oct 6;7:411. doi: 10.3389/fimmu.2016.00411. eCollection 2016. Front Immunol. 2016. PMID: 27766099 Free PMC article.
-
Induction and maintenance of IL-4 expression are regulated differently by the 3' enhancer in CD4 T cells.J Immunol. 2011 Mar 1;186(5):2792-9. doi: 10.4049/jimmunol.1003353. Epub 2011 Jan 31. J Immunol. 2011. PMID: 21282512 Free PMC article.
-
Development of innate CD4+ and CD8+ T cells in Itk-deficient mice is regulated by distinct pathways.J Immunol. 2014 Jul 15;193(2):688-99. doi: 10.4049/jimmunol.1302059. Epub 2014 Jun 18. J Immunol. 2014. PMID: 24943215 Free PMC article.
-
Transcriptional regulation of the NKT cell lineage.Curr Opin Immunol. 2013 Apr;25(2):161-7. doi: 10.1016/j.coi.2013.01.003. Epub 2013 Feb 9. Curr Opin Immunol. 2013. PMID: 23402834 Free PMC article. Review.
-
The transcription factor BCL-6 controls early development of innate-like T cells.Nat Immunol. 2020 Sep;21(9):1058-1069. doi: 10.1038/s41590-020-0737-y. Epub 2020 Jul 27. Nat Immunol. 2020. PMID: 32719520 Free PMC article.
References
-
- Alonzo ES, Gottschalk RA, Das J, Egawa T, Hobbs RM, Pandolfi PP, Pereira P, Nichols KE, Koretzky GA, Jordan MS, Sant’angelo DB. Development of Promyelocytic Zinc Finger and ThPOK-Expressing Innate {gamma}{delta} T Cells Is Controlled by Strength of TCR Signaling and Id3 3. J Immunol 2009 - PMC - PubMed
-
- Atherly LO, Lucas JA, Felices M, Yin CC, Reiner SL, Berg LJ. The Tec family tyrosine kinases Itk and Rlk regulate the development of conventional CD8+ T cells. Immunity. 2006;25:79–91. - PubMed
-
- Azuara V, Levraud JP, Lembezat MP, Pereira P. A novel subset of adult gamma delta thymocytes that secretes a distinct pattern of cytokines and expresses a very restricted T cell receptor repertoire. Eur J Immunol. 1997;27:544–553. - PubMed
-
- Bain G, Cravatt CB, Loomans C, Alberola-Ila J, Hedrick SM, Murre C. Regulation of the helix-loop-helix proteins, E2A and Id3, by the Ras-ERK MAPK cascade. Nat Immunol. 2001;2:165–171. - PubMed
-
- Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

