Immune-related zinc finger gene ZFAT is an essential transcriptional regulator for hematopoietic differentiation in blood islands
- PMID: 20660741
- PMCID: PMC2922575
- DOI: 10.1073/pnas.1002494107
Immune-related zinc finger gene ZFAT is an essential transcriptional regulator for hematopoietic differentiation in blood islands
Abstract
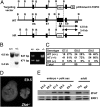
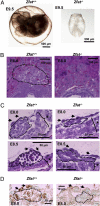
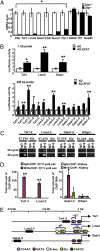
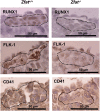
TAL1 plays pivotal roles in vascular and hematopoietic developments through the complex with LMO2 and GATA1. Hemangioblasts, which have a differentiation potential for both endothelial and hematopoietic lineages, arise in the primitive streak and migrate into the yolk sac to form blood islands, where primitive hematopoiesis occurs. ZFAT (a zinc-finger gene in autoimmune thyroid disease susceptibility region/an immune-related transcriptional regulator containing 18 C(2)H(2)-type zinc-finger domains and one AT-hook) was originally identified as an immune-related transcriptional regulator containing 18 C(2)H(2)-type zinc-finger domains and one AT-hook, and is highly conserved among species. ZFAT is thought to be a critical transcription factor involved in immune-regulation and apoptosis; however, developmental roles for ZFAT remain unknown. Here we show that Zfat-deficient (Zfat(-/-)) mice are embryonic-lethal, with impaired differentiation of hematopoietic progenitor cells in blood islands, where ZFAT is exactly expressed. Expression levels of Tal1, Lmo2, and Gata1 in Zfat(-/-) yolk sacs are much reduced compared with those of wild-type mice, and ChIP-PCR analysis revealed that ZFAT binds promoter regions for these genes in vivo. Furthermore, profound reduction in TAL1, LMO2, and GATA1 protein expressions are observed in Zfat(-/-) blood islands. Taken together, these results suggest that ZFAT is indispensable for mouse embryonic development and functions as a critical transcription factor for primitive hematopoiesis through direct-regulation of Tal1, Lmo2, and Gata1. Elucidation of ZFAT functions in hematopoiesis might lead to a better understanding of transcriptional networks in differentiation and cellular programs of hematopoietic lineage and provide useful information for applied medicine in stem cell therapy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
ZFAT is a critical molecule for cell survival in mouse embryonic fibroblasts.Cell Mol Biol Lett. 2011 Mar;16(1):89-100. doi: 10.2478/s11658-010-0041-1. Epub 2010 Dec 27. Cell Mol Biol Lett. 2011. PMID: 21225468 Free PMC article.
-
Essential roles for Cdx in murine primitive hematopoiesis.Dev Biol. 2017 Feb 15;422(2):115-124. doi: 10.1016/j.ydbio.2017.01.002. Epub 2017 Jan 6. Dev Biol. 2017. PMID: 28065741
-
Structure of the leukemia oncogene LMO2: implications for the assembly of a hematopoietic transcription factor complex.Blood. 2011 Feb 17;117(7):2146-56. doi: 10.1182/blood-2010-07-293357. Epub 2010 Nov 12. Blood. 2011. PMID: 21076045
-
Roles of ZFAT in haematopoiesis, angiogenesis and cancer development.Anticancer Res. 2013 Jul;33(7):2833-7. Anticancer Res. 2013. PMID: 23780967 Review.
-
Regulation of hemangioblast development.Ann N Y Acad Sci. 2001 Jun;938:96-107; discussion 108. doi: 10.1111/j.1749-6632.2001.tb03578.x. Ann N Y Acad Sci. 2001. PMID: 11458531 Review.
Cited by
-
Solution structures of the DNA-binding domains of immune-related zinc-finger protein ZFAT.J Struct Funct Genomics. 2015 Jun;16(2):55-65. doi: 10.1007/s10969-015-9196-3. Epub 2015 Mar 24. J Struct Funct Genomics. 2015. PMID: 25801860 Free PMC article.
-
ZFAT is a critical molecule for cell survival in mouse embryonic fibroblasts.Cell Mol Biol Lett. 2011 Mar;16(1):89-100. doi: 10.2478/s11658-010-0041-1. Epub 2010 Dec 27. Cell Mol Biol Lett. 2011. PMID: 21225468 Free PMC article.
-
Bioinformatic screening of autoimmune disease genes and protein structure prediction with FAMS for drug discovery.Protein Pept Lett. 2014;21(8):828-39. doi: 10.2174/09298665113209990052. Protein Pept Lett. 2014. PMID: 23855671 Free PMC article.
-
ZFAT binds to centromeres to control noncoding RNA transcription through the KAT2B-H4K8ac-BRD4 axis.Nucleic Acids Res. 2020 Nov 4;48(19):10848-10866. doi: 10.1093/nar/gkaa815. Nucleic Acids Res. 2020. PMID: 32997115 Free PMC article.
-
The Nuclear Zinc Finger Protein Zfat Maintains FoxO1 Protein Levels in Peripheral T Cells by Regulating the Activities of Autophagy and the Akt Signaling Pathway.J Biol Chem. 2016 Jul 15;291(29):15282-91. doi: 10.1074/jbc.M116.723734. Epub 2016 May 20. J Biol Chem. 2016. PMID: 27226588 Free PMC article.
References
-
- Gläsker S, et al. Hemangioblastomas share protein expression with embryonal hemangioblast progenitor cell. Cancer Res. 2006;66:4167–4172. - PubMed
-
- Palis J, Yoder MC. Yolk-sac hematopoiesis: The first blood cells of mouse and man. Exp Hematol. 2001;29:927–936. - PubMed
-
- Huber TL, Kouskoff V, Fehling HJ, Palis J, Keller G. Haemangioblast commitment is initiated in the primitive streak of the mouse embryo. Nature. 2004;432:625–630. - PubMed
-
- Coultas L, Chawengsaksophak K, Rossant J. Endothelial cells and VEGF in vascular development. Nature. 2005;438:937–945. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

