XBP-1 couples endoplasmic reticulum stress to augmented IFN-beta induction via a cis-acting enhancer in macrophages
- PMID: 20660350
- PMCID: PMC2916979
- DOI: 10.4049/jimmunol.0903052
XBP-1 couples endoplasmic reticulum stress to augmented IFN-beta induction via a cis-acting enhancer in macrophages
Abstract
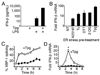
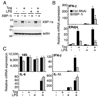
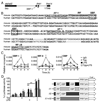
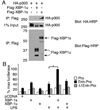
Perturbation of the endoplasmic reticulum (ER) results in a conserved stress response called the unfolded protein response (UPR). Macrophages undergoing a UPR respond to LPS with log-fold increased production of IFN-beta, a cytokine with diverse roles in innate and adaptive immunity. In this study, we found that thapsigargin-induced ER stress augmented recruitment of IFN regulatory factor-3, CREB binding protein/p300, and transcriptional machinery to the murine ifnb1 promoter during LPS stimulation. Although full synergistic IFN-beta production requires X-box binding protein 1 (XBP-1), this UPR-regulated transcription factor did not appreciably bind the ifnb1 promoter. However, XBP-1 bound a conserved site 6.1 kb downstream of ifnb1, along with IFN regulatory factor-3 and CREB binding protein only during concomitant UPR and LPS stimulation. XBP-1 physically associates with p300, suggesting a mechanism of multimolecular assembly at the +6.1 kb site. Luciferase reporter assays provide evidence this +6 kb region functions as an XBP-1-dependent enhancer of ifnb1 promoter activity. Thus, this study identifies a novel role for a UPR-dependent transcription factor in the regulation of an inflammatory cytokine. Our findings have broader mechanistic implications for the pathogenesis of diseases involving ER stress and type I IFN, including viral infection, ischemia-reperfusion injury, protein misfolding, and inflammatory diseases.
Figures


Similar articles
-
Endoplasmic reticulum stress and the unfolded protein response are linked to synergistic IFN-beta induction via X-box binding protein 1.Eur J Immunol. 2008 May;38(5):1194-203. doi: 10.1002/eji.200737882. Eur J Immunol. 2008. PMID: 18412159 Free PMC article.
-
The integrated endoplasmic reticulum stress response in Leishmania amazonensis macrophage infection: the role of X-box binding protein 1 transcription factor.FASEB J. 2016 Apr;30(4):1557-65. doi: 10.1096/fj.15-281550. Epub 2015 Dec 17. FASEB J. 2016. PMID: 26678450 Free PMC article.
-
Endoplasmic reticulum stress-induced IRE1α activation mediates cross-talk of GSK-3β and XBP-1 to regulate inflammatory cytokine production.J Immunol. 2015 May 1;194(9):4498-506. doi: 10.4049/jimmunol.1401399. Epub 2015 Mar 27. J Immunol. 2015. PMID: 25821218 Free PMC article.
-
The X-box binding protein-1 transcription factor is required for plasma cell differentiation and the unfolded protein response.Immunol Rev. 2003 Aug;194:29-38. doi: 10.1034/j.1600-065x.2003.00057.x. Immunol Rev. 2003. PMID: 12846805 Review.
-
The transcription factor X-box binding protein-1 in neurodegenerative diseases.Mol Neurodegener. 2014 Sep 12;9:35. doi: 10.1186/1750-1326-9-35. Mol Neurodegener. 2014. PMID: 25216759 Free PMC article. Review.
Cited by
-
The UPR and lung disease.Semin Immunopathol. 2013 May;35(3):293-306. doi: 10.1007/s00281-013-0368-6. Epub 2013 Mar 28. Semin Immunopathol. 2013. PMID: 23536202 Review.
-
Macrophage mTORC1 disruption reduces inflammation and insulin resistance in obese mice.Diabetologia. 2014 Nov;57(11):2393-404. doi: 10.1007/s00125-014-3350-5. Epub 2014 Aug 14. Diabetologia. 2014. PMID: 25120095
-
Roles of XBP1s in Transcriptional Regulation of Target Genes.Biomedicines. 2021 Jul 8;9(7):791. doi: 10.3390/biomedicines9070791. Biomedicines. 2021. PMID: 34356855 Free PMC article. Review.
-
Endoplasmic reticulum stress-activated glycogen synthase kinase 3β aggravates liver inflammation and hepatotoxicity in mice with acute liver failure.Inflammation. 2015;38(3):1151-65. doi: 10.1007/s10753-014-0080-2. Inflammation. 2015. PMID: 25630719
-
The ataxia telangiectasia mutated kinase pathway regulates IL-23 expression by human dendritic cells.J Immunol. 2013 Apr 1;190(7):3246-55. doi: 10.4049/jimmunol.1201484. Epub 2013 Mar 4. J Immunol. 2013. PMID: 23460736 Free PMC article.
References
-
- Theofilopoulos AN, Baccala R, Beutler B, Kono DH. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu Rev Immunol. 2005;23:307–336. - PubMed
-
- Honda K, Taniguchi T. Toll-like receptor signaling and IRF transcription factors. IUBMB Life. 2006;58:290–295. - PubMed
-
- Beg AA. Endogenous ligands of Toll-like receptors: implications for regulating inflammatory and immune responses. Trends Immunol. 2002;23:509–512. - PubMed
-
- Thomas KE, Galligan CL, Newman RD, Fish EN, Vogel SN. Contribution of interferon-beta to the murine macrophage response to the toll-like receptor 4 agonist, lipopolysaccharide. J Biol Chem. 2006;281:31119–31130. - PubMed
-
- Mancuso G, Midiri A, Biondo C, Beninati C, Zummo S, Galbo R, Tomasello F, Gambuzza M, Macri G, Ruggeri A, Leanderson T, Teti G. Type I IFN signaling is crucial for host resistance against different species of pathogenic bacteria. J Immunol. 2007;178:3126–3133. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K08 AI081045-01/AI/NIAID NIH HHS/United States
- UL1 RR025011-03/RR/NCRR NIH HHS/United States
- UL1 RR025011-03S2/RR/NCRR NIH HHS/United States
- UL1 RR025011/RR/NCRR NIH HHS/United States
- KL2 UL1RR025011/RR/NCRR NIH HHS/United States
- UL1 RR025011-03S1/RR/NCRR NIH HHS/United States
- R01 DK082582/DK/NIDDK NIH HHS/United States
- R01 DK082582-02/DK/NIDDK NIH HHS/United States
- UL1 RR025011-02/RR/NCRR NIH HHS/United States
- UL1 RR025011-01/RR/NCRR NIH HHS/United States
- UL1 RR025011-04/RR/NCRR NIH HHS/United States
- KL2 K08-AI081045/AI/NIAID NIH HHS/United States
- K08 AI081045-02/AI/NIAID NIH HHS/United States
- K08 AI081045/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

