Nature, position, and frequency of mutations made in a single cycle of HIV-1 replication
- PMID: 20660205
- PMCID: PMC2937799
- DOI: 10.1128/JVI.00915-10
Nature, position, and frequency of mutations made in a single cycle of HIV-1 replication
Abstract
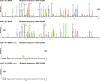
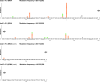
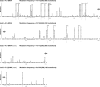
There is considerable HIV-1 variation in patients. The extent of the variation is due to the high rate of viral replication, the high viral load, and the errors made during viral replication. Mutations can arise from errors made either by host DNA-dependent RNA polymerase II or by HIV-1 reverse transcriptase (RT), but the relative contributions of these two enzymes to the mutation rate are unknown. In addition, mutations in RT can affect its fidelity, but the effect of mutations in RT on the nature of the mutations that arise in vivo is poorly understood. We have developed an efficient system, based on existing technology, to analyze the mutations that arise in an HIV-1 vector in a single cycle of replication. A lacZalpha reporter gene is used to identify viral DNAs that contain mutations which are analyzed by DNA sequencing. The forward mutation rate in this system is 1.4 x 10(-5) mutations/bp/cycle, equivalent to the retroviral average. This rate is about 3-fold lower than previously reported for HIV-1 in vivo and is much lower than what has been reported for purified HIV-1 RT in vitro. Although the mutation rate was not affected by the orientation of lacZalpha, the sites favored for mutations (hot spots) in lacZalpha depended on which strand of lacZalpha was present in the viral RNA. The pattern of hot spots seen in lacZalpha in vivo did not match any of the published data obtained when purified RT was used to copy lacZalpha in vitro.
Figures

Similar articles
-
Mutations in HIV-1 reverse transcriptase affect the errors made in a single cycle of viral replication.J Virol. 2014 Jul;88(13):7589-601. doi: 10.1128/JVI.00302-14. Epub 2014 Apr 23. J Virol. 2014. PMID: 24760888 Free PMC article.
-
Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase.J Virol. 1995 Aug;69(8):5087-94. doi: 10.1128/JVI.69.8.5087-5094.1995. J Virol. 1995. PMID: 7541846 Free PMC article.
-
Mutational analysis of HIV-1 long terminal repeats to explore the relative contribution of reverse transcriptase and RNA polymerase II to viral mutagenesis.J Biol Chem. 2002 Oct 11;277(41):38053-61. doi: 10.1074/jbc.M204774200. Epub 2002 Jul 31. J Biol Chem. 2002. PMID: 12151398
-
Molecular basis of fidelity of DNA synthesis and nucleotide specificity of retroviral reverse transcriptases.Prog Nucleic Acid Res Mol Biol. 2002;71:91-147. doi: 10.1016/s0079-6603(02)71042-8. Prog Nucleic Acid Res Mol Biol. 2002. PMID: 12102562 Review.
-
Nucleoside-analog resistance mutations in HIV-1 reverse transcriptase and their influence on polymerase fidelity and viral mutation rates.Int J Biochem Cell Biol. 2004 Sep;36(9):1716-34. doi: 10.1016/j.biocel.2004.02.025. Int J Biochem Cell Biol. 2004. PMID: 15183340 Review.
Cited by
-
Biological Barriers for Drug Delivery and Development of Innovative Therapeutic Approaches in HIV, Pancreatic Cancer, and Hemophilia A/B.Pharmaceutics. 2024 Sep 13;16(9):1207. doi: 10.3390/pharmaceutics16091207. Pharmaceutics. 2024. PMID: 39339243 Free PMC article. Review.
-
Chronic HIV Transcription, Translation, and Persistent Inflammation.Viruses. 2024 May 9;16(5):751. doi: 10.3390/v16050751. Viruses. 2024. PMID: 38793632 Free PMC article. Review.
-
Combating antimicrobial resistance in malaria, HIV and tuberculosis.Nat Rev Drug Discov. 2024 Jun;23(6):461-479. doi: 10.1038/s41573-024-00933-4. Epub 2024 May 15. Nat Rev Drug Discov. 2024. PMID: 38750260 Review.
-
The Question of HIV Vaccine: Why Is a Solution Not Yet Available?J Immunol Res. 2024 Apr 8;2024:2147912. doi: 10.1155/2024/2147912. eCollection 2024. J Immunol Res. 2024. PMID: 38628675 Free PMC article. Review.
-
Modeling resistance to the broadly neutralizing antibody PGT121 in people living with HIV-1.PLoS Comput Biol. 2024 Mar 29;20(3):e1011518. doi: 10.1371/journal.pcbi.1011518. eCollection 2024 Mar. PLoS Comput Biol. 2024. PMID: 38551976 Free PMC article.
References
-
- Avidan, O., M. E. Meer, I. Oz, and A. Hizi. 2002. The processivity and fidelity of DNA synthesis exhibited by the reverse transcriptase of bovine leukemia virus. Eur. J. Biochem. 269:859-867. - PubMed
-
- Bakhanashvili, M., and A. Hizi. 1993. Fidelity of DNA synthesis exhibited in vitro by the reverse transcriptase of the lentivirus equine infectious anemia virus. Biochemistry 32:7559-7567. - PubMed
-
- Bebenek, K., J. Abbotts, J. D. Roberts, S. H. Wilson, and T. A. Kunkel. 1989. Specificity and mechanism of error-prone replication by human immunodeficiency virus-1 reverse transcriptase. J. Biol. Chem. 264:16948-16956. - PubMed
-
- Bebenek, K., J. Abbotts, S. H. Wilson, and T. A. Kunkel. 1993. Error-prone polymerization by HIV-1 reverse transcriptase. Contribution of template-primer misalignment, miscoding, and termination probability to mutational hot spots. J. Biol. Chem. 268:10324-10334. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

