A systems approach to mapping transcriptional networks controlling surfactant homeostasis
- PMID: 20659319
- PMCID: PMC3091648
- DOI: 10.1186/1471-2164-11-451
A systems approach to mapping transcriptional networks controlling surfactant homeostasis
Abstract
Background: Pulmonary surfactant is required for lung function at birth and throughout life. Lung lipid and surfactant homeostasis requires regulation among multi-tiered processes, coordinating the synthesis of surfactant proteins and lipids, their assembly, trafficking, and storage in type II cells of the lung. The mechanisms regulating these interrelated processes are largely unknown.
Results: We integrated mRNA microarray data with array independent knowledge using Gene Ontology (GO) similarity analysis, promoter motif searching, protein interaction and literature mining to elucidate genetic networks regulating lipid related biological processes in lung. A Transcription factor (TF)-target gene (TG) similarity matrix was generated by integrating data from different analytic methods. A scoring function was built to rank the likely TF-TG pairs. Using this strategy, we identified and verified critical components of a transcriptional network directing lipogenesis, lipid trafficking and surfactant homeostasis in the mouse lung.
Conclusions: Within the transcriptional network, SREBP, CEBPA, FOXA2, ETSF, GATA6 and IRF1 were identified as regulatory hubs displaying high connectivity. SREBP, FOXA2 and CEBPA together form a common core regulatory module that controls surfactant lipid homeostasis. The core module cooperates with other factors to regulate lipid metabolism and transport, cell growth and development, cell death and cell mediated immune response. Coordinated interactions of the TFs influence surfactant homeostasis and regulate lung function at birth.
Figures

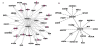
Similar articles
-
Gene expression and biological processes influenced by deletion of Stat3 in pulmonary type II epithelial cells.BMC Genomics. 2007 Dec 10;8:455. doi: 10.1186/1471-2164-8-455. BMC Genomics. 2007. PMID: 18070348 Free PMC article.
-
C/EBP{alpha} is required for pulmonary cytoprotection during hyperoxia.Am J Physiol Lung Cell Mol Physiol. 2009 Aug;297(2):L286-98. doi: 10.1152/ajplung.00094.2009. Epub 2009 May 22. Am J Physiol Lung Cell Mol Physiol. 2009. PMID: 19465518 Free PMC article.
-
Sterol response element binding protein and thyroid transcription factor-1 (Nkx2.1) regulate Abca3 gene expression.Am J Physiol Lung Cell Mol Physiol. 2007 Dec;293(6):L1395-405. doi: 10.1152/ajplung.00275.2007. Epub 2007 Sep 21. Am J Physiol Lung Cell Mol Physiol. 2007. PMID: 17890326
-
Pulmonary surfactants and their role in pathophysiology of lung disorders.Indian J Exp Biol. 2013 Jan;51(1):5-22. Indian J Exp Biol. 2013. PMID: 23441475 Review.
-
Review: The intersection of surfactant homeostasis and innate host defense of the lung: lessons from newborn infants.Innate Immun. 2010 Jun;16(3):138-42. doi: 10.1177/1753425910366879. Epub 2010 Mar 29. Innate Immun. 2010. PMID: 20351134 Review.
Cited by
-
LKB1 drives stasis and C/EBP-mediated reprogramming to an alveolar type II fate in lung cancer.Nat Commun. 2022 Feb 28;13(1):1090. doi: 10.1038/s41467-022-28619-8. Nat Commun. 2022. PMID: 35228570 Free PMC article.
-
Building and Regenerating the Lung Cell by Cell.Physiol Rev. 2019 Jan 1;99(1):513-554. doi: 10.1152/physrev.00001.2018. Physiol Rev. 2019. PMID: 30427276 Free PMC article. Review.
-
XAGE-1b expression is associated with the diagnosis and early recurrence of hepatocellular carcinoma.Mol Clin Oncol. 2014 Nov;2(6):1155-1159. doi: 10.3892/mco.2014.336. Epub 2014 Jul 4. Mol Clin Oncol. 2014. PMID: 25279215 Free PMC article.
-
Systems biology approaches to identify developmental bases for lung diseases.Pediatr Res. 2013 Apr;73(4 Pt 2):514-22. doi: 10.1038/pr.2013.7. Epub 2013 Jan 11. Pediatr Res. 2013. PMID: 23314295 Free PMC article. Review.
-
Population differences in transcript-regulator expression quantitative trait loci.PLoS One. 2012;7(3):e34286. doi: 10.1371/journal.pone.0034286. Epub 2012 Mar 27. PLoS One. 2012. PMID: 22479588 Free PMC article.
References
-
- Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, Goldstein JL. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci USA. 2003;100(21):12027–12032. doi: 10.1073/pnas.1534923100. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

