Splice-site mutations cause Rrp6-mediated nuclear retention of the unspliced RNAs and transcriptional down-regulation of the splicing-defective genes
- PMID: 20634951
- PMCID: PMC2902512
- DOI: 10.1371/journal.pone.0011540
Splice-site mutations cause Rrp6-mediated nuclear retention of the unspliced RNAs and transcriptional down-regulation of the splicing-defective genes
Abstract
Background: Eukaryotic cells have developed surveillance mechanisms to prevent the expression of aberrant transcripts. An early surveillance checkpoint acts at the transcription site and prevents the release of mRNAs that carry processing defects. The exosome subunit Rrp6 is required for this checkpoint in Saccharomyces cerevisiae, but it is not known whether Rrp6 also plays a role in mRNA surveillance in higher eukaryotes.
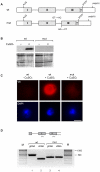
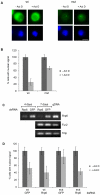
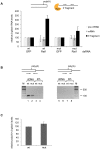
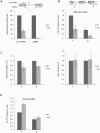
Methodology/principal findings: We have developed an in vivo system to study nuclear mRNA surveillance in Drosophila melanogaster. We have produced S2 cells that express a human beta-globin gene with mutated splice sites in intron 2 (mut beta-globin). The transcripts encoded by the mut beta-globin gene are normally spliced at intron 1 but retain intron 2. The levels of the mut beta-globin transcripts are much lower than those of wild type (wt) ss-globin mRNAs transcribed from the same promoter. We have compared the expression of the mut and wt beta-globin genes to investigate the mechanisms that down-regulate the production of defective mRNAs. Both wt and mut beta-globin transcripts are processed at the 3', but the mut beta-globin transcripts are less efficiently cleaved than the wt transcripts. Moreover, the mut beta-globin transcripts are less efficiently released from the transcription site, as shown by FISH, and this defect is restored by depletion of Rrp6 by RNAi. Furthermore, transcription of the mut beta-globin gene is significantly impaired as revealed by ChIP experiments that measure the association of the RNA polymerase II with the transcribed genes. We have also shown that the mut beta-globin gene shows reduced levels of H3K4me3.
Conclusions/significance: Our results show that there are at least two surveillance responses that operate cotranscriptionally in insect cells and probably in all metazoans. One response requires Rrp6 and results in the inefficient release of defective mRNAs from the transcription site. The other response acts at the transcription level and reduces the synthesis of the defective transcripts through a mechanism that involves histone modifications.
Conflict of interest statement
Figures
Similar articles
-
Rrp6 is recruited to transcribed genes and accompanies the spliced mRNA to the nuclear pore.RNA. 2012 Aug;18(8):1466-74. doi: 10.1261/rna.032045.111. Epub 2012 Jun 28. RNA. 2012. PMID: 22745224 Free PMC article.
-
A link between nuclear RNA surveillance, the human exosome and RNA polymerase II transcriptional termination.Nucleic Acids Res. 2010 Dec;38(22):8015-26. doi: 10.1093/nar/gkq703. Epub 2010 Aug 10. Nucleic Acids Res. 2010. PMID: 20699273 Free PMC article.
-
The exosome component Rrp6 is required for RNA polymerase II termination at specific targets of the Nrd1-Nab3 pathway.PLoS Genet. 2015 Feb 13;11(2):e1004999. doi: 10.1371/journal.pgen.1004999. eCollection 2015. PLoS Genet. 2015. PMID: 25680078 Free PMC article.
-
Control of human beta-globin mRNA stability and its impact on beta-thalassemia phenotype.Haematologica. 2011 Jun;96(6):905-13. doi: 10.3324/haematol.2010.039206. Epub 2011 Feb 28. Haematologica. 2011. PMID: 21357703 Free PMC article. Review.
-
Rrp6: Integrated roles in nuclear RNA metabolism and transcription termination.Wiley Interdiscip Rev RNA. 2016 Jan-Feb;7(1):91-104. doi: 10.1002/wrna.1317. Epub 2015 Nov 26. Wiley Interdiscip Rev RNA. 2016. PMID: 26612606 Free PMC article. Review.
Cited by
-
Splicing-coupled 3' end formation requires a terminal splice acceptor site, but not intron excision.Nucleic Acids Res. 2013 Aug;41(14):7101-14. doi: 10.1093/nar/gkt446. Epub 2013 May 28. Nucleic Acids Res. 2013. PMID: 23716637 Free PMC article.
-
Global analysis of the nuclear processing of transcripts with unspliced U12-type introns by the exosome.Nucleic Acids Res. 2014 Jun;42(11):7358-69. doi: 10.1093/nar/gku391. Epub 2014 May 21. Nucleic Acids Res. 2014. PMID: 24848017 Free PMC article.
-
Where splicing joins chromatin.Nucleus. 2011 May-Jun;2(3):182-8. doi: 10.4161/nucl.2.3.15876. Nucleus. 2011. PMID: 21818411 Free PMC article. Review.
-
Proofreading and spellchecking: a two-tier strategy for pre-mRNA splicing quality control.RNA. 2011 Mar;17(3):383-9. doi: 10.1261/rna.2454711. Epub 2011 Jan 4. RNA. 2011. PMID: 21205840 Free PMC article. Review.
-
Coupling and coordination in gene expression processes with pre-mRNA splicing.Int J Mol Sci. 2015 Mar 11;16(3):5682-96. doi: 10.3390/ijms16035682. Int J Mol Sci. 2015. PMID: 25768347 Free PMC article. Review.
References
-
- Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3:195–205. - PubMed
-
- Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. - PubMed
-
- Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. - PubMed
-
- Neugebauer KM. On the importance of being co-transcriptional. J Cell Sci. 2002;115:3865–3871. - PubMed
-
- Allemand E, Batsché E, Muchardt C. Splicing, transcription, and chromatin: a ménage à trois. Curr Opin Genet Dev. 2008;18:145–151. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

