A chromatin-remodeling protein is a component of fission yeast mediator
- PMID: 20622008
- PMCID: PMC2943280
- DOI: 10.1074/jbc.M110.153858
A chromatin-remodeling protein is a component of fission yeast mediator
Abstract
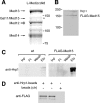
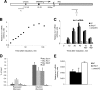
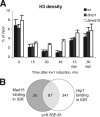
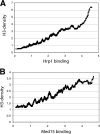
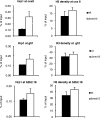
The multiprotein Mediator complex is an important regulator of RNA polymerase II-dependent genes in eukaryotic cells. In contrast to the situation in many other eukaryotes, the conserved Med15 protein is not a stable component of Mediator isolated from fission yeast. We here demonstrate that Med15 exists in a protein complex together with Hrp1, a CHD1 ATP-dependent chromatin-remodeling protein. The Med15-Hrp1 subcomplex is not a component of the core Mediator complex but can interact with the L-Mediator conformation. Deletion of med15(+) and hrp1(+) causes very similar effects on global steady-state levels of mRNA, and genome-wide analyses demonstrate that Med15 associates with a distinct subset of Hrp1-bound gene promoters. Our findings therefore indicate that Mediator may directly influence histone density at regulated promoters.
Figures
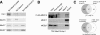
Similar articles
-
Identification of noncoding transcripts from within CENP-A chromatin at fission yeast centromeres.J Biol Chem. 2011 Jul 1;286(26):23600-7. doi: 10.1074/jbc.M111.228510. Epub 2011 Apr 28. J Biol Chem. 2011. PMID: 21531710 Free PMC article.
-
Genome-wide characterization of Mediator recruitment, function, and regulation.Transcription. 2017 May 27;8(3):169-174. doi: 10.1080/21541264.2017.1291082. Epub 2017 Feb 8. Transcription. 2017. PMID: 28301289 Free PMC article. Review.
-
Topoisomerase I regulates open chromatin and controls gene expression in vivo.EMBO J. 2010 Jul 7;29(13):2126-34. doi: 10.1038/emboj.2010.109. Epub 2010 Jun 4. EMBO J. 2010. PMID: 20526281 Free PMC article.
-
Chd1 chromatin remodelers maintain nucleosome organization and repress cryptic transcription.EMBO Rep. 2012 Nov 6;13(11):997-1003. doi: 10.1038/embor.2012.146. Epub 2012 Oct 2. EMBO Rep. 2012. PMID: 23032292 Free PMC article.
-
Epigenetic Regulation of Chromatin States in Schizosaccharomyces pombe.Cold Spring Harb Perspect Biol. 2015 Jul 1;7(7):a018770. doi: 10.1101/cshperspect.a018770. Cold Spring Harb Perspect Biol. 2015. PMID: 26134317 Free PMC article. Review.
Cited by
-
Post-translational modifications of histones that influence nucleosome dynamics.Chem Rev. 2015 Mar 25;115(6):2274-95. doi: 10.1021/cr500350x. Epub 2014 Nov 26. Chem Rev. 2015. PMID: 25424540 Free PMC article. Review. No abstract available.
-
Cyclin C influences the timing of mitosis in fission yeast.Mol Biol Cell. 2017 Jul 1;28(13):1738-1744. doi: 10.1091/mbc.E16-11-0787. Epub 2017 May 17. Mol Biol Cell. 2017. PMID: 28515143 Free PMC article.
-
A key role for Chd1 in histone H3 dynamics at the 3' ends of long genes in yeast.PLoS Genet. 2012;8(7):e1002811. doi: 10.1371/journal.pgen.1002811. Epub 2012 Jul 12. PLoS Genet. 2012. PMID: 22807688 Free PMC article.
-
A functional portrait of Med7 and the mediator complex in Candida albicans.PLoS Genet. 2014 Nov 6;10(11):e1004770. doi: 10.1371/journal.pgen.1004770. eCollection 2014 Nov. PLoS Genet. 2014. PMID: 25375174 Free PMC article.
-
Transcription-coupled recruitment of human CHD1 and CHD2 influences chromatin accessibility and histone H3 and H3.3 occupancy at active chromatin regions.Epigenetics Chromatin. 2015 Jan 15;8(1):4. doi: 10.1186/1756-8935-8-4. eCollection 2015. Epigenetics Chromatin. 2015. PMID: 25621013 Free PMC article.
References
-
- Holstege F. C., Jennings E. G., Wyrick J. J., Lee T. I., Hengartner C. J., Green M. R., Golub T. R., Lander E. S., Young R. A. (1998) Cell 95, 717–728 - PubMed
-
- Kornberg R. D. (2005) Trends Biochem. Sci. 30, 235–239 - PubMed
-
- Björklund S., Gustafsson C. M. (2005) Trends Biochem. Sci. 30, 240–244 - PubMed
-
- Kim Y. J., Björklund S., Li Y., Sayre M. H., Kornberg R. D. (1994) Cell 77, 599–608 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

