DNA uptake sequence-mediated enhancement of transformation in Neisseria gonorrhoeae is strain dependent
- PMID: 20601472
- PMCID: PMC2937394
- DOI: 10.1128/JB.00442-10
DNA uptake sequence-mediated enhancement of transformation in Neisseria gonorrhoeae is strain dependent
Abstract
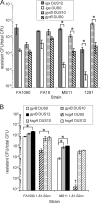
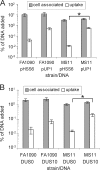
Natural transformation is the main means of horizontal genetic exchange in the obligate human pathogen Neisseria gonorrhoeae. Neisseria spp. have been shown to preferentially take up and transform their own DNA by recognizing the nonpalindromic 10- or 12-nucleotide sequence 5'-ATGCCGTCTGAA-3' (additional semiconserved nucleotides are underlined), termed the DNA uptake sequence (DUS10 or DUS12). Here we investigated the effects of the DUS on transformation and DNA uptake for several laboratory strains of N. gonorrhoeae. We found that all strains showed efficient transformation of DUS containing DNA (DUS10 and DUS12) but that the level of transformation with DNA lacking a DUS (DUS0) was variable in different strains. The DUS-enhanced transformation was 20-fold in two strains, FA1090 and FA19, but was approximately 150-fold in strains MS11 and 1291. All strains tested provide some level of DUS0 transformation, and DUS0 transformation was type IV pilus dependent. Competition with plasmid DNA revealed that transformation of MS11 was enhanced by the addition of excess plasmid DNA containing a DUS while FA1090 transformation was competitively inhibited. Although FA1090 was able to mediate much more efficient transformation of DNA lacking a DUS than was MS11, DNA uptake experiments showed similar levels of uptake of DNA containing and lacking a DUS in FA1090 and MS11. Finally, DNA uptake was competitively inhibited in both FA1090 and MS11. Taken together, our data indicate that the role of the DUS during DNA transformation is variable between strains of N. gonorrhoeae and may influence multiple steps during transformation.
Figures

Similar articles
-
Genetic transformation of Neisseria gonorrhoeae shows a strand preference.FEMS Microbiol Lett. 2012 Sep;334(1):44-8. doi: 10.1111/j.1574-6968.2012.02612.x. Epub 2012 Jul 3. FEMS Microbiol Lett. 2012. PMID: 22676068 Free PMC article.
-
Pilin antigenic variation occurs independently of the RecBCD pathway in Neisseria gonorrhoeae.J Bacteriol. 2009 Sep;191(18):5613-21. doi: 10.1128/JB.00535-09. Epub 2009 Jul 10. J Bacteriol. 2009. PMID: 19592592 Free PMC article.
-
PilC of Neisseria meningitidis is involved in class II pilus formation and restores pilus assembly, natural transformation competence and adherence to epithelial cells in PilC-deficient gonococci.Mol Microbiol. 1997 Mar;23(5):879-92. doi: 10.1046/j.1365-2958.1997.2631630.x. Mol Microbiol. 1997. PMID: 9076726
-
Transformation competence and type-4 pilus biogenesis in Neisseria gonorrhoeae--a review.Gene. 1997 Jun 11;192(1):125-34. doi: 10.1016/s0378-1119(97)00038-3. Gene. 1997. PMID: 9224882 Review.
-
Competence for natural transformation in Neisseria gonorrhoeae: a model system for studies of horizontal gene transfer.APMIS Suppl. 1998;84:56-61. doi: 10.1111/j.1600-0463.1998.tb05649.x. APMIS Suppl. 1998. PMID: 9850683 Review.
Cited by
-
Mobile DNA in the pathogenic Neisseria.Microbiol Spectr. 2015 Feb;3(3):MDNA3-0015-2014. doi: 10.1128/microbiolspec.MDNA3-0015-2014. Microbiol Spectr. 2015. PMID: 25866700 Free PMC article.
-
Antimicrobial Resistance Profiles of Human Commensal Neisseria Species.Antibiotics (Basel). 2021 May 6;10(5):538. doi: 10.3390/antibiotics10050538. Antibiotics (Basel). 2021. PMID: 34066576 Free PMC article.
-
Neisseria gonorrhoeae NGO2105 Is an Autotransporter Protein Involved in Adhesion to Human Cervical Epithelial Cells and in vivo Colonization.Front Microbiol. 2020 Jun 25;11:1395. doi: 10.3389/fmicb.2020.01395. eCollection 2020. Front Microbiol. 2020. PMID: 32670242 Free PMC article.
-
The Pilin N-terminal Domain Maintains Neisseria gonorrhoeae Transformation Competence during Pilus Phase Variation.PLoS Genet. 2016 May 23;12(5):e1006069. doi: 10.1371/journal.pgen.1006069. eCollection 2016 May. PLoS Genet. 2016. PMID: 27213957 Free PMC article.
-
Analysis of Pilin Antigenic Variation in Neisseria meningitidis by Next-Generation Sequencing.J Bacteriol. 2018 Oct 23;200(22):e00465-18. doi: 10.1128/JB.00465-18. Print 2018 Nov 15. J Bacteriol. 2018. PMID: 30181126 Free PMC article.
References
-
- Aas, F. E., C. Lovold, and M. Koomey. 2002. An inhibitor of DNA binding and uptake events dictates the proficiency of genetic transformation in Neisseria gonorrhoeae: mechanism of action and links to type IV pilus expression. Mol. Microbiol. 46:1441-1450. - PubMed
-
- Aas, F. E., M. Wolfgang, S. Frye, S. Dunham, C. Lovold, and M. Koomey. 2002. Competence for natural transformation in Neisseria gonorrhoeae: components of DNA binding and uptake linked to type IV pilus expression. Mol. Microbiol. 46:749-760. - PubMed
-
- Anonymous. 2007. Update to CDC's Sexually Transmitted Diseases Treatment Guidelines, 2006: fluoroquinolones no longer recommended for treatment of gonococcal infections. MMWR Morb. Mortal. Wkly. Rep. 56:332-336. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

