Transcription through the HIV-1 nucleosomes: effects of the PBAF complex in Tat activated transcription
- PMID: 20599239
- PMCID: PMC2923259
- DOI: 10.1016/j.virol.2010.06.009
Transcription through the HIV-1 nucleosomes: effects of the PBAF complex in Tat activated transcription
Abstract
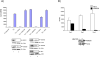
The SWI/SNF complex remodels nucleosomes, allowing RNA Polymerase II access to the HIV-1 proviral DNA. It has not been determined which SWI/SNF complex (BAF or PBAF) remodels nucleosomes at the transcription start site. These complexes differ in only three subunits and determining which subunit(s) is required could explain the regulation of Tat activated transcription. We show that PBAF is required for chromatin remodeling at the nuc-1 start site and transcriptional elongation. We find that Baf200 is required to ensure activation at the LTR level and for viral production. Interestingly, the BAF complex was observed on the LTR whereas PBAF was present on both LTR and Env regions. We found that Tat activated transcription facilitates removal of histones H2A and H2B at the LTR, and that the FACT complex may be responsible for their removal. Finally, the BAF complex may play an important role in regulating splicing of the HIV-1 genome.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures


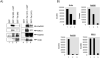

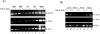
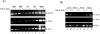


Similar articles
-
Effect of SWI/SNF chromatin remodeling complex on HIV-1 Tat activated transcription.Retrovirology. 2006 Aug 7;3:48. doi: 10.1186/1742-4690-3-48. Retrovirology. 2006. PMID: 16893449 Free PMC article.
-
Repressive LTR nucleosome positioning by the BAF complex is required for HIV latency.PLoS Biol. 2011 Nov;9(11):e1001206. doi: 10.1371/journal.pbio.1001206. Epub 2011 Nov 29. PLoS Biol. 2011. PMID: 22140357 Free PMC article.
-
Varying modulation of HIV-1 LTR activity by Baf complexes.J Mol Biol. 2011 Aug 19;411(3):581-96. doi: 10.1016/j.jmb.2011.06.001. Epub 2011 Jun 15. J Mol Biol. 2011. PMID: 21699904 Free PMC article.
-
Chromatin remodeling and modification during HIV-1 Tat-activated transcription.Curr HIV Res. 2003 Jul;1(3):343-62. doi: 10.2174/1570162033485186. Curr HIV Res. 2003. PMID: 15046258 Review.
-
Chromatin-associated regulation of HIV-1 transcription: implications for the development of therapeutic strategies.Subcell Biochem. 2007;41:371-96. Subcell Biochem. 2007. PMID: 17484137 Review.
Cited by
-
The Tat inhibitor didehydro-cortistatin A suppresses SIV replication and reactivation.FASEB J. 2019 Jul;33(7):8280-8293. doi: 10.1096/fj.201801165R. Epub 2019 Apr 25. FASEB J. 2019. PMID: 31021670 Free PMC article.
-
The Short Isoform of BRD4 Promotes HIV-1 Latency by Engaging Repressive SWI/SNF Chromatin-Remodeling Complexes.Mol Cell. 2017 Sep 21;67(6):1001-1012.e6. doi: 10.1016/j.molcel.2017.07.025. Epub 2017 Aug 24. Mol Cell. 2017. PMID: 28844864 Free PMC article.
-
Exosomes derived from HIV-1-infected cells contain trans-activation response element RNA.J Biol Chem. 2013 Jul 5;288(27):20014-33. doi: 10.1074/jbc.M112.438895. Epub 2013 May 9. J Biol Chem. 2013. PMID: 23661700 Free PMC article.
-
Block-And-Lock: New Horizons for a Cure for HIV-1.Viruses. 2020 Dec 15;12(12):1443. doi: 10.3390/v12121443. Viruses. 2020. PMID: 33334019 Free PMC article. Review.
-
Role of Host Factors on the Regulation of Tat-Mediated HIV-1 Transcription.Curr Pharm Des. 2017;23(28):4079-4090. doi: 10.2174/1381612823666170622104355. Curr Pharm Des. 2017. PMID: 28641539 Free PMC article. Review.
References
-
- Batsche E, Yaniv M, Muchardt C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat Struct Mol Biol. 2006;13(1):22–9. - PubMed
-
- Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301(5636):1090–3. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

