Biotin regulates the expression of holocarboxylase synthetase in the miR-539 pathway in HEK-293 cells
- PMID: 20592104
- PMCID: PMC2924595
- DOI: 10.3945/jn.110.126359
Biotin regulates the expression of holocarboxylase synthetase in the miR-539 pathway in HEK-293 cells
Abstract
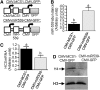
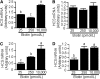
Holocarboxylase synthetase (HCS) catalyzes the covalent binding of biotin to carboxylases and histones. In mammals, the expression of HCS depends on biotin, but the mechanism of regulation is unknown. Here we tested the hypothesis that microRNA (miR) plays a role in the regulation of the HCS gene. Human embryonic kidney cells were used as the primary model, but cell lines from other tissues and primary human cells were also tested. In silico searches revealed an evolutionary conserved binding site for miR-539 in the 3 prime -untranslated region (3 prime -UTR) of HCS mRNA. Transgenic cells and reporter gene constructs were used to demonstrate that miR-539 decreases the expression of HCS at the level of transcription rather than translation; these findings were corroborated in nontransgenic cells. When miR-539 was overexpressed in transgenic cells, the abundance of both HCS and biotinylated histones decreased. The abundance of miR-539 was tissue dependent: fibroblasts gt kidney cells gt intestinal cells gt lymphoid cells. Dose-response studies revealed that the abundance of miR-539 was significantly higher at physiological concentrations of biotin than both biotin-deficient and biotin-supplemented media in all cell lines tested. In kidney cells, the expression of HCS was lower in cells in physiological medium than in deficient and supplemented medium. In contrast, in fibroblasts, lymphoid cells, and intestinal cells, there was no apparent link between miR-539 abundance and HCS expression, suggesting that factors other than miR-539 also contribute to the regulation of HCS expression in some tissues. Collectively, the results of this study suggest that miR-539 is among the factors sensing biotin and regulating HCS.
Conflict of interest statement
Author disclosures: B. Bao, R. Rodriguez-Melendez, S. S. K. Wijeratne and J. Zempleni, no conflicts of interest.
Figures


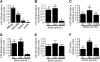
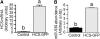
Similar articles
-
Cytosine methylation in miR-153 gene promoters increases the expression of holocarboxylase synthetase, thereby increasing the abundance of histone H4 biotinylation marks in HEK-293 human kidney cells.J Nutr Biochem. 2012 Jun;23(6):635-9. doi: 10.1016/j.jnutbio.2011.03.007. Epub 2011 Jul 20. J Nutr Biochem. 2012. PMID: 21764280 Free PMC article.
-
Holocarboxylase synthetase is an obligate participant in biotin-mediated regulation of its own expression and of biotin-dependent carboxylases mRNA levels in human cells.Proc Natl Acad Sci U S A. 2002 Apr 16;99(8):5325-30. doi: 10.1073/pnas.082097699. Proc Natl Acad Sci U S A. 2002. PMID: 11959985 Free PMC article.
-
Biotin regulates the genetic expression of holocarboxylase synthetase and mitochondrial carboxylases in rats.J Nutr. 2001 Jul;131(7):1909-13. doi: 10.1093/jn/131.7.1909. J Nutr. 2001. PMID: 11435506
-
Holocarboxylase Synthetase: A Moonlighting Transcriptional Coregulator of Gene Expression and a Cytosolic Regulator of Biotin Utilization.Annu Rev Nutr. 2017 Aug 21;37:207-223. doi: 10.1146/annurev-nutr-042617-104653. Epub 2017 May 31. Annu Rev Nutr. 2017. PMID: 28564555 Review.
-
Molecular genetics of biotin metabolism: old vitamin, new science.J Nutr Biochem. 2005 Jul;16(7):428-31. doi: 10.1016/j.jnutbio.2005.03.020. J Nutr Biochem. 2005. PMID: 15992684 Review.
Cited by
-
Holocarboxylase synthetase interacts physically with nuclear receptor co-repressor, histone deacetylase 1 and a novel splicing variant of histone deacetylase 1 to repress repeats.Biochem J. 2014 Aug 1;461(3):477-86. doi: 10.1042/BJ20131208. Biochem J. 2014. PMID: 24840043 Free PMC article.
-
The role of holocarboxylase synthetase in genome stability is mediated partly by epigenomic synergies between methylation and biotinylation events.Epigenetics. 2011 Jul;6(7):892-4. doi: 10.4161/epi.6.7.15544. Epub 2011 Jul 1. Epigenetics. 2011. PMID: 21555910 Free PMC article.
-
Post-transcriptional dysregulation by miRNAs is implicated in the pathogenesis of gastrointestinal stromal tumor [GIST].PLoS One. 2013 May 24;8(5):e64102. doi: 10.1371/journal.pone.0064102. Print 2013. PLoS One. 2013. PMID: 23717541 Free PMC article.
-
Breast Milk Supply of MicroRNA Associated with Leptin and Adiponectin Is Affected by Maternal Overweight/Obesity and Influences Infancy BMI.Nutrients. 2019 Oct 28;11(11):2589. doi: 10.3390/nu11112589. Nutrients. 2019. PMID: 31661820 Free PMC article.
-
Niacin in pharmacological doses alters microRNA expression in skeletal muscle of obese Zucker rats.PLoS One. 2014 May 21;9(5):e98313. doi: 10.1371/journal.pone.0098313. eCollection 2014. PLoS One. 2014. PMID: 24847987 Free PMC article.
References
-
- Manthey KC, Griffin JB, Zempleni J. Biotin supply affects expression of biotin transporters, biotinylation of carboxylases, and metabolism of interleukin-2 in Jurkat cells. J Nutr. 2002;132:887–92 - PubMed
-
- Mock DM, Johnson SB, Holman RT. Effects of biotin deficiency on serum fatty acid composition: Evidence for abnormalities in humans. J Nutr. 1988;118:342–8 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

