Distribution and paralogue specificity of mammalian deSUMOylating enzymes
- PMID: 20590526
- PMCID: PMC3749516
- DOI: 10.1042/BJ20100504
Distribution and paralogue specificity of mammalian deSUMOylating enzymes
Abstract
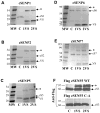
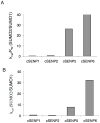
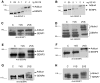
The covalent attachment of SUMO (small ubiquitin-like protein modifier) to target proteins results in modifications in their activity, binding interactions, localization or half-life. The reversal of this modification is catalysed by SENPs (SUMO-specific processing proteases). Mammals contain four SUMO paralogues and six SENP enzymes. In the present paper, we describe a systematic analysis of human SENPs, integrating estimates of relative selectivity for SUMO1 and SUMO2, and kinetic measurements of recombinant C-terminal cSENPs (SENP catalytic domains). We first characterized the reaction of each endogenous SENP and cSENPs with HA-SUMO-VS [HA (haemagglutinin)-tagged SUMO-vinyl sulfones], active-site-directed irreversible inhibitors of SENPs. We found that all cSENPs and endogenous SENP1 react with both SUMO paralogues, whereas all other endogenous SENPs in mammalian cells and tissues display high selectivity for SUMO2-VS. To obtain more quantitative data, the kinetic properties of purified cSENPs were determined using SUMO1- or SUMO2-AMC (7-amino-4-methylcoumarin) as substrate. All enzymes bind their respective substrates with high affinity. cSENP1 and cSENP2 process either SUMO substrate with similar affinity and catalytic efficiency; cSENP5 and cSENP6 show marked catalytic specificity for SUMO2 as measured by Km and kcat, whereas cSENP7 works only on SUMO2. Compared with cSENPs, recombinant full-length SENP1 and SENP2 show differences in SUMO selectivity, indicating that paralogue specificity is influenced by the presence of the variable N-terminal domain of each SENP. Our data suggest that SUMO2 metabolism is more dynamic than that of SUMO1 since most SENPs display a marked preference for SUMO2.
Figures

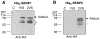

Similar articles
-
Evaluation of the activity and substrate specificity of the human SENP family of SUMO proteases.Biochim Biophys Acta. 2016 Jan;1863(1):139-47. doi: 10.1016/j.bbamcr.2015.10.020. Epub 2015 Oct 30. Biochim Biophys Acta. 2016. PMID: 26522917
-
Senp1 is essential for desumoylating Sumo1-modified proteins but dispensable for Sumo2 and Sumo3 deconjugation in the mouse embryo.Cell Rep. 2013 May 30;3(5):1640-50. doi: 10.1016/j.celrep.2013.04.016. Epub 2013 May 16. Cell Rep. 2013. PMID: 23684609 Free PMC article.
-
DeSUMOylating enzymes--SENPs.IUBMB Life. 2008 Nov;60(11):734-42. doi: 10.1002/iub.113. IUBMB Life. 2008. PMID: 18666185 Review.
-
Swapping small ubiquitin-like modifier (SUMO) isoform specificity of SUMO proteases SENP6 and SENP7.J Biol Chem. 2011 Oct 14;286(41):36142-36151. doi: 10.1074/jbc.M111.268847. Epub 2011 Aug 30. J Biol Chem. 2011. PMID: 21878624 Free PMC article.
-
Paralogue-Specific Roles of SUMO1 and SUMO2/3 in Protein Quality Control and Associated Diseases.Cells. 2023 Dec 20;13(1):8. doi: 10.3390/cells13010008. Cells. 2023. PMID: 38201212 Free PMC article. Review.
Cited by
-
A snapshot of the Ixodes scapularis degradome.Gene. 2011 Aug 15;482(1-2):78-93. doi: 10.1016/j.gene.2011.04.008. Epub 2011 Apr 28. Gene. 2011. PMID: 21596113 Free PMC article.
-
Identification and characterization of a new chemotype of noncovalent SENP inhibitors.ACS Chem Biol. 2013 Jul 19;8(7):1435-41. doi: 10.1021/cb400177q. Epub 2013 May 1. ACS Chem Biol. 2013. PMID: 23614497 Free PMC article.
-
SENP3-mediated deSUMOylation of dynamin-related protein 1 promotes cell death following ischaemia.EMBO J. 2013 May 29;32(11):1514-28. doi: 10.1038/emboj.2013.65. Epub 2013 Mar 22. EMBO J. 2013. PMID: 23524851 Free PMC article.
-
SUMO deconjugation is required for arsenic-triggered ubiquitylation of PML.Sci Signal. 2015 Jun 9;8(380):ra56. doi: 10.1126/scisignal.aaa3929. Sci Signal. 2015. PMID: 26060329 Free PMC article.
-
Polypharmacology of small molecules targeting the ubiquitin-proteasome and ubiquitin-like systems.Oncotarget. 2015;6(12):9646-56. doi: 10.18632/oncotarget.3917. Oncotarget. 2015. PMID: 25991664 Free PMC article. Review.
References
-
- Seeler JS, Dejean A. Nuclear and unclear functions of SUMO. Nat Rev Mol Cell Biol. 2003;4:690–699. - PubMed
-
- Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. - PubMed
-
- Tatham MH, Jaffray E, Vaughan OA, Desterro JM, Botting CH, Naismith JH, Hay RT. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem. 2001;276:35368–35374. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

