Quantitative analysis of the mechanism of endocytic actin patch assembly and disassembly in fission yeast
- PMID: 20587778
- PMCID: PMC2921122
- DOI: 10.1091/mbc.E10-02-0157
Quantitative analysis of the mechanism of endocytic actin patch assembly and disassembly in fission yeast
Abstract
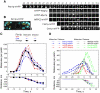
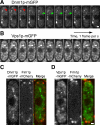
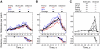
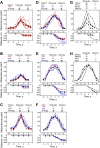
We used quantitative confocal microscopy to measure the numbers of 16 proteins tagged with fluorescent proteins during assembly and disassembly of endocytic actin patches in fission yeast. The peak numbers of each molecule that accumulate in patches varied <30-50% between individual patches. The pathway begins with accumulation of 30-40 clathrin molecules, sufficient to build a hemisphere at the tip of a plasma membrane invagination. Thereafter precisely timed waves of proteins reach characteristic peak numbers: endocytic adaptor proteins (approximately 120 End4p and approximately 230 Pan1p), activators of Arp2/3 complex (approximately 200 Wsp1p and approximately 340 Myo1p) and approximately 300 Arp2/3 complexes just ahead of a burst of actin assembly into short, capped and highly cross-linked filaments (approximately 7000 actins, approximately 200 capping proteins, and approximately 900 fimbrins). Coronin arrives last as all other components disperse upon patch internalization and movement over approximately 10 s. Patch internalization occurs without recruitment of dynamins. Mathematical modeling, described in the accompanying paper (Berro et al., 2010, MBoC 21: 2803-2813), shows that the dendritic nucleation hypothesis can account for the time course of actin assembly into a branched network of several hundred filaments 100-200 nm long and that patch disassembly requires actin filament fragmentation in addition to depolymerization from the ends.
Figures


Similar articles
-
Mathematical modeling of endocytic actin patch kinetics in fission yeast: disassembly requires release of actin filament fragments.Mol Biol Cell. 2010 Aug 15;21(16):2905-15. doi: 10.1091/mbc.E10-06-0494. Epub 2010 Jun 29. Mol Biol Cell. 2010. PMID: 20587776 Free PMC article.
-
Distinct roles for F-BAR proteins Cdc15p and Bzz1p in actin polymerization at sites of endocytosis in fission yeast.Curr Biol. 2011 Sep 13;21(17):1450-9. doi: 10.1016/j.cub.2011.07.046. Epub 2011 Sep 1. Curr Biol. 2011. PMID: 21885283 Free PMC article.
-
High-speed superresolution imaging of the proteins in fission yeast clathrin-mediated endocytic actin patches.Mol Biol Cell. 2018 Feb 1;29(3):295-303. doi: 10.1091/mbc.E17-06-0415. Epub 2017 Dec 6. Mol Biol Cell. 2018. PMID: 29212877 Free PMC article.
-
Pan1p: an actin director of endocytosis in yeast.Int J Biochem Cell Biol. 2007;39(10):1760-4. doi: 10.1016/j.biocel.2006.12.001. Epub 2006 Dec 21. Int J Biochem Cell Biol. 2007. PMID: 17303466 Review.
-
Coupling actin dynamics and membrane dynamics during endocytosis.Curr Opin Cell Biol. 2002 Feb;14(1):76-81. doi: 10.1016/s0955-0674(01)00297-6. Curr Opin Cell Biol. 2002. PMID: 11792548 Review.
Cited by
-
Cytosolic concentrations of actin binding proteins and the implications for in vivo F-actin turnover.J Cell Biol. 2023 Dec 4;222(12):e202306036. doi: 10.1083/jcb.202306036. Epub 2023 Oct 6. J Cell Biol. 2023. PMID: 37801069 Free PMC article.
-
Regarding the amazing choreography of clathrin coats.PLoS Biol. 2011 Mar;9(3):e1001037. doi: 10.1371/journal.pbio.1001037. Epub 2011 Mar 22. PLoS Biol. 2011. PMID: 21445329 Free PMC article. No abstract available.
-
Internetwork competition for monomers governs actin cytoskeleton organization.Nat Rev Mol Cell Biol. 2016 Dec;17(12):799-810. doi: 10.1038/nrm.2016.106. Epub 2016 Sep 14. Nat Rev Mol Cell Biol. 2016. PMID: 27625321 Free PMC article.
-
Actin and endocytosis in budding yeast.Genetics. 2015 Feb;199(2):315-58. doi: 10.1534/genetics.112.145540. Genetics. 2015. PMID: 25657349 Free PMC article. Review.
-
Identification of an ATP-controlled allosteric switch that controls actin filament nucleation by Arp2/3 complex.Nat Commun. 2016 Jul 15;7:12226. doi: 10.1038/ncomms12226. Nat Commun. 2016. PMID: 27417392 Free PMC article.
References
-
- Bahler J., Wu J. Q., Longtine M. S., Shah N. G., McKenzie A., 3rd, Steever A. B., Wach A., Philippsen P., Pringle J. R. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. - PubMed
-
- Fotin A., Cheng Y., Sliz P., Grigorieff N., Harrison S. C., Kirchhausen T., Walz T. Molecular model for a complete clathrin lattice from electron cryomicroscopy. Nature. 2004;432:573–579. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

