The minimal autoinhibited unit of the guanine nucleotide exchange factor intersectin
- PMID: 20585582
- PMCID: PMC2892021
- DOI: 10.1371/journal.pone.0011291
The minimal autoinhibited unit of the guanine nucleotide exchange factor intersectin
Abstract
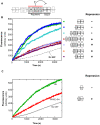
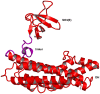
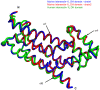
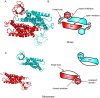
Intersectin-1L is a member of the Dbl homology (DH) domain guanine nucleotide exchange factors (GEF) which control Rho-family GTPase signaling. Intersectin-1L is a GEF that is specific for Cdc42. It plays an important role in endocytosis, and is regulated by several partners including the actin regulator N-WASP. Intact intersectin-1L shows low Cdc42 exchange activity, although the isolated catalytic DH domain shows high activity. This finding suggests that the molecule is autoinhibited. To investigate the mechanism of autoinhibition we have constructed a series of domain deletions. We find that the five SH3 domains of intersectin are important for autoinhibition, with the fifth domain (SH3(E)) being sufficient for the bulk of the autoinhibitory effect. This SH3 domain appears to primarily interact with the DH domain. We have determined the crystal structure of the SH3(E)-DH domain construct, which shows a domain swapped arrangement in which the SH3 from one monomer interacts with the DH domain of the other monomer. Analytical ultracentrifugation and gel filtration, however, show that under biochemical concentrations, the construct is fully monomeric. Thus we propose that the actual autoinhibited structure contains the related intramolecular SH3(E)-DH interaction. We propose a model in which this intramolecular interaction may block or distort the GTPase binding region of the DH domain.
Conflict of interest statement
Figures


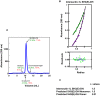
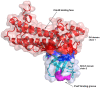
Similar articles
-
Autoinhibition of GEF activity in Intersectin 1 is mediated by the short SH3-DH domain linker.Protein Sci. 2010 Nov;19(11):2164-74. doi: 10.1002/pro.500. Protein Sci. 2010. PMID: 20842712 Free PMC article.
-
Intersectin 1L guanine nucleotide exchange activity is regulated by adjacent src homology 3 domains that are also involved in endocytosis.Mol Biol Cell. 2003 Apr;14(4):1624-37. doi: 10.1091/mbc.e02-08-0494. Mol Biol Cell. 2003. PMID: 12686614 Free PMC article.
-
A Cdc42 mutant specifically activated by intersectin.Biochemistry. 2005 Oct 11;44(40):13282-90. doi: 10.1021/bi050591b. Biochemistry. 2005. PMID: 16201754
-
The Rho guanine nucleotide exchange factors Intersectin 1L and β-Pix control calcium-regulated exocytosis in neuroendocrine PC12 cells.Cell Mol Neurobiol. 2010 Nov;30(8):1327-33. doi: 10.1007/s10571-010-9580-2. Epub 2010 Nov 19. Cell Mol Neurobiol. 2010. PMID: 21088884 Review.
-
The guanine nucleotide exchange factor Tiam1: a Janus-faced molecule in cellular signaling.Cell Signal. 2014 Mar;26(3):483-91. doi: 10.1016/j.cellsig.2013.11.034. Epub 2013 Dec 2. Cell Signal. 2014. PMID: 24308970 Review.
Cited by
-
Regulating Cdc42 and Its Signaling Pathways in Cancer: Small Molecules and MicroRNA as New Treatment Candidates.Molecules. 2018 Mar 29;23(4):787. doi: 10.3390/molecules23040787. Molecules. 2018. PMID: 29596304 Free PMC article. Review.
-
An optimized growth medium for increased recombinant protein secretion titer via the type III secretion system.Microb Cell Fact. 2021 Feb 15;20(1):44. doi: 10.1186/s12934-021-01536-z. Microb Cell Fact. 2021. PMID: 33588857 Free PMC article.
-
Structure of the Rho-specific guanine nucleotide-exchange factor Xpln.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012 Dec 1;68(Pt 12):1455-9. doi: 10.1107/S1744309112045265. Epub 2012 Nov 19. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012. PMID: 23192023 Free PMC article.
-
Structural basis for autoinhibition of the guanine nucleotide exchange factor FARP2.Structure. 2013 Mar 5;21(3):355-64. doi: 10.1016/j.str.2013.01.001. Epub 2013 Jan 31. Structure. 2013. PMID: 23375260 Free PMC article.
-
Autoinhibition of GEF activity in Intersectin 1 is mediated by the short SH3-DH domain linker.Protein Sci. 2010 Nov;19(11):2164-74. doi: 10.1002/pro.500. Protein Sci. 2010. PMID: 20842712 Free PMC article.
References
-
- Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–180. - PubMed
-
- Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. - PubMed
-
- Hoffman GR, Cerione RA. Signaling to the Rho GTPases: networking with the DH domain. FEBS Lett. 2002;513:85–91. - PubMed
-
- Van Aelst L, D'Souza-Schorey C. Rho GTPases and signaling networks. Genes Dev. 1997;11:2295–2322. - PubMed
-
- Mackay DJ, Hall A. Rho GTPases. J Biol Chem. 1998;273:20685–20688. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

