The NF-kappaB p50:p50:HDAC-1 repressor complex orchestrates transcriptional inhibition of multiple pro-inflammatory genes
- PMID: 20579762
- PMCID: PMC3098379
- DOI: 10.1016/j.jhep.2010.03.025
The NF-kappaB p50:p50:HDAC-1 repressor complex orchestrates transcriptional inhibition of multiple pro-inflammatory genes
Abstract
Background & aims: The pro-inflammatory functions of NF-kappaB must be tightly regulated to prevent inappropriate tissue damage and remodelling caused by activated inflammatory and wound-healing cells. The p50 subunit of NF-kappaB is emerging as an important repressor of immune and inflammatory responses, but by mechanisms that are poorly defined. This study aims to delineate p50 target genes in activated hepatic stellate cells and to outline mechanisms utilised in their repression.
Methods: Hepatic stellate cells were isolated from nfkb1(p50)-deficient or Wt mice and gene expression compared using microarray. Target genes were verified by qRT-PCR and p50-mediated HDAC-1 recruitment to the target genes demonstrated using chromatin immunoprecipitation.
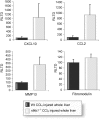
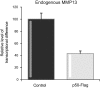
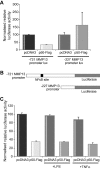
Results: We identify p50 as transcriptional repressor of multiple pro-inflammatory genes including Ccl2, Cxcl10, Gm-csf, and Mmp-13. These genes are over-expressed in nfkb1(p50)-deficient mice suffering from chronic hepatitis and in fibrogenic/inflammatory hepatic stellate cells isolated from nfkb1(-/-) liver. We identify Mmp-13 as a bona-fide target gene for p50 and demonstrate that p50 is required for recruitment of the transcriptional repressor histone deacetylase (HDAC)-1 to kappaB sites in the Mmp-13 promoter. Chromatin immunoprecipitations identified binding of HDAC-1 to specific regulatory regions of the Ccl2, Cxcl10, Gm-csf genes that contain predicted kappaB binding motifs. Recruitment of HDAC-1 to these genes was not observed in nfkb1(-/-) cells suggesting a requirement for p50 in a manner similar to that described for Mmp-13.
Conclusions: Recruitment of HDAC-1 to inflammatory genes provides a widespread mechanism to explain the immunosuppressive properties of p50.
Copyright 2010 European Association for the Study of the Liver. Published by Elsevier B.V. All rights reserved.
Figures

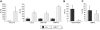


Similar articles
-
NF-kappa B p50 regulates C/EBP alpha expression and inflammatory cytokine-induced neutrophil production.J Immunol. 2009 May 1;182(9):5757-62. doi: 10.4049/jimmunol.0803861. J Immunol. 2009. PMID: 19380823 Free PMC article.
-
HDAC1 interacts with the p50 NF-?B subunit via its nuclear localization sequence to constrain inflammatory gene expression.Biochim Biophys Acta Gene Regul Mech. 2018 Oct;1861(10):962-970. doi: 10.1016/j.bbagrm.2018.09.001. Epub 2018 Sep 10. Biochim Biophys Acta Gene Regul Mech. 2018. PMID: 30496041
-
Induction of matrix metalloproteinase-1 gene transcription by tumour necrosis factor alpha via the p50/p50 homodimer of nuclear factor-kappa B in activated human hepatic stellate cells.Liver Int. 2008 Dec;28(10):1418-25. doi: 10.1111/j.1478-3231.2008.01883.x. Liver Int. 2008. PMID: 19055644
-
NFKB1: a suppressor of inflammation, ageing and cancer.FEBS J. 2016 May;283(10):1812-22. doi: 10.1111/febs.13627. Epub 2016 Jan 13. FEBS J. 2016. PMID: 26663363 Review.
-
Role of the NF-κB system in context-specific tuning of the inflammatory gene response.Curr Opin Immunol. 2021 Feb;68:21-27. doi: 10.1016/j.coi.2020.08.005. Epub 2020 Sep 6. Curr Opin Immunol. 2021. PMID: 32898750 Review.
Cited by
-
Innate immune response of human alveolar macrophages during influenza A infection.PLoS One. 2012;7(3):e29879. doi: 10.1371/journal.pone.0029879. Epub 2012 Mar 2. PLoS One. 2012. PMID: 22396727 Free PMC article.
-
Hsp90 inhibition increases SOCS3 transcript and regulates migration and cell death in chronic lymphocytic leukemia.Oncotarget. 2016 May 10;7(19):28684-96. doi: 10.18632/oncotarget.8760. Oncotarget. 2016. PMID: 27107422 Free PMC article.
-
NFKB1 and Cancer: Friend or Foe?Cells. 2018 Sep 7;7(9):133. doi: 10.3390/cells7090133. Cells. 2018. PMID: 30205516 Free PMC article. Review.
-
Epigenetic reprogramming in liver fibrosis and cancer.Adv Drug Deliv Rev. 2017 Nov 1;121:124-132. doi: 10.1016/j.addr.2017.10.011. Epub 2017 Oct 25. Adv Drug Deliv Rev. 2017. PMID: 29079534 Free PMC article. Review.
-
Moderate Exercise Inhibits Age-Related Inflammation, Liver Steatosis, Senescence, and Tumorigenesis.J Immunol. 2021 Feb 15;206(4):904-916. doi: 10.4049/jimmunol.2001022. Epub 2021 Jan 13. J Immunol. 2021. PMID: 33441438 Free PMC article.
References
-
- Adcock I., Evans P.C. Resolution of inflammatory responses: a brief introduction. Biochem Soc Trans. 2007;35:261–262. - PubMed
-
- Bondeson J., Brennan F., Foxwell B., Feldmann M. Effective adenoviral transfer of IkappaBalpha into human fibroblasts and chondrosarcoma cells reveals that the induction of matrix metalloproteinases and proinflammatory cytokines is nuclear factor-kappaB dependent. J Rheumatol. 2000;27:2078–2089. - PubMed
-
- Chen L.W., Egan L., Li Z.W., Greten F.R., Kagnoff M.F., Karin M. The two faces of IKK and NF-kappaB inhibition: prevention of systemic inflammation but increased local injury following intestinal ischemia–reperfusion. Nat Med. 2003;9:575–581. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

