Distal regions of the human IFNG locus direct cell type-specific expression
- PMID: 20574006
- PMCID: PMC2923829
- DOI: 10.4049/jimmunol.1000124
Distal regions of the human IFNG locus direct cell type-specific expression
Abstract
Genes, such as IFNG, which are expressed in multiple cell lineages of the immune system, may employ a common set of regulatory elements to direct transcription in multiple cell types or individual regulatory elements to direct expression in individual cell lineages. By employing a bacterial artificial chromosome transgenic system, we demonstrate that IFNG employs unique regulatory elements to achieve lineage-specific transcriptional control. Specifically, a one 1-kb element 30 kb upstream of IFNG activates transcription in T cells and NKT cells but not in NK cells. This distal regulatory element is a Runx3 binding site in Th1 cells and is needed for RNA polymerase II recruitment to IFNG, but it is not absolutely required for histone acetylation of the IFNG locus. These results support a model whereby IFNG uses cis-regulatory elements with cell type-restricted function.
Conflict of interest statement
The authors have no financial conflicts of interest.
Figures
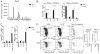


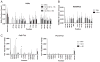
Similar articles
-
Diverse functions of distal regulatory elements at the IFNG locus.J Immunol. 2012 Feb 15;188(4):1726-33. doi: 10.4049/jimmunol.1102879. Epub 2012 Jan 13. J Immunol. 2012. PMID: 22246629 Free PMC article.
-
A distal conserved sequence element controls Ifng gene expression by T cells and NK cells.Immunity. 2006 Nov;25(5):717-29. doi: 10.1016/j.immuni.2006.09.007. Epub 2006 Oct 26. Immunity. 2006. PMID: 17070076
-
Deletion of a conserved cis-element in the Ifng locus highlights the role of acute histone acetylation in modulating inducible gene transcription.PLoS Genet. 2014 Jan;10(1):e1003969. doi: 10.1371/journal.pgen.1003969. Epub 2014 Jan 9. PLoS Genet. 2014. PMID: 24415943 Free PMC article.
-
Regulation of interferon-gamma during innate and adaptive immune responses.Adv Immunol. 2007;96:41-101. doi: 10.1016/S0065-2776(07)96002-2. Adv Immunol. 2007. PMID: 17981204 Review.
-
Regulation of the Ifng locus in the context of T-lineage specification and plasticity.Immunol Rev. 2010 Nov;238(1):216-32. doi: 10.1111/j.1600-065X.2010.00961.x. Immunol Rev. 2010. PMID: 20969595 Free PMC article. Review.
Cited by
-
Regulation of the Th1 genomic locus from Ifng through Tmevpg1 by T-bet.J Immunol. 2014 Oct 15;193(8):3959-65. doi: 10.4049/jimmunol.1401099. Epub 2014 Sep 15. J Immunol. 2014. PMID: 25225667 Free PMC article.
-
Lineage-specific adjacent IFNG and IL26 genes share a common distal enhancer element.Genes Immun. 2012 Sep;13(6):481-8. doi: 10.1038/gene.2012.22. Epub 2012 May 24. Genes Immun. 2012. PMID: 22622197 Free PMC article.
-
Proximal and Distal Regions of Pathogenic Th17 Related Chromatin Loci Are Sequentially Accessible During Pathogenicity of Th17.Front Immunol. 2022 Apr 19;13:864314. doi: 10.3389/fimmu.2022.864314. eCollection 2022. Front Immunol. 2022. PMID: 35514969 Free PMC article.
-
Transcriptional and epigenetic regulation of T-helper lineage specification.Immunol Rev. 2014 Sep;261(1):62-83. doi: 10.1111/imr.12204. Immunol Rev. 2014. PMID: 25123277 Free PMC article. Review.
-
Cutting edge: influence of Tmevpg1, a long intergenic noncoding RNA, on the expression of Ifng by Th1 cells.J Immunol. 2012 Sep 1;189(5):2084-8. doi: 10.4049/jimmunol.1200774. Epub 2012 Jul 30. J Immunol. 2012. PMID: 22851706 Free PMC article.
References
-
- Woolfe A, Elgar G. Organization of conserved elements near key developmental regulators in vertebrate genomes. Adv Genet. 2008;61:307–338. - PubMed
-
- Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol. 2009;9:91–105. - PubMed
-
- Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–669. - PubMed
-
- Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, Zediak VP, Banica M, DiCioccio CB, Gross DA, Mao CA, et al. Control of effector CD8+ T cell function by the transcription factor eomesodermin. Science. 2003;302:1041–1043. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R56 AI044924/AI/NIAID NIH HHS/United States
- R01 AI044924-09A1/AI/NIAID NIH HHS/United States
- P30 CA68485/CA/NCI NIH HHS/United States
- R01 AI044924-01A2/AI/NIAID NIH HHS/United States
- R56 AI044924-09/AI/NIAID NIH HHS/United States
- T32 HL069765/HL/NHLBI NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- HL069765/HL/NHLBI NIH HHS/United States
- P01 AI056296/AI/NIAID NIH HHS/United States
- DK058404/DK/NIDDK NIH HHS/United States
- CA48126/CA/NCI NIH HHS/United States
- AI56296/AI/NIAID NIH HHS/United States
- P30 DK058404/DK/NIDDK NIH HHS/United States
- AI44924/AI/NIAID NIH HHS/United States
- R01 AI044924/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

