Spatial regulation of Golgi phosphatidylinositol-4-phosphate is required for enzyme localization and glycosylation fidelity
- PMID: 20573065
- PMCID: PMC2921771
- DOI: 10.1111/j.1600-0854.2010.01092.x
Spatial regulation of Golgi phosphatidylinositol-4-phosphate is required for enzyme localization and glycosylation fidelity
Abstract
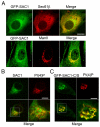
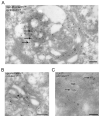
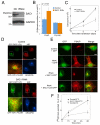
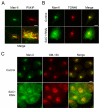
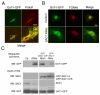
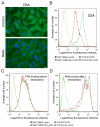
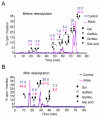
The enrichment of phosphatidylinositol-4-phosphate (PI(4)P) at the trans Golgi network (TGN) is instrumental for proper protein and lipid sorting, yet how the restricted distribution of PI(4)P is achieved remains unknown. Here, we show that lipid phosphatase Suppressor of actin mutations 1 (SAC1) is crucial for the spatial regulation of Golgi PI(4)P. Ultrastructural analysis revealed that SAC1 is predominantly located at cisternal Golgi membranes but is absent from the TGN, thus confining PI(4)P to the TGN. RNAi-mediated knockdown of SAC1 caused changes in Golgi morphology and mislocalization of Golgi enzymes. Enzymes involved in glycan processing such as mannosidase-II (Man-II) and N-acetylglucosamine transferase-I (GnT-I) redistributed to aberrant intracellular structures and to the cell surface in SAC1 knockdown cells. SAC1 depletion also induced a unique pattern of Golgi-specific defects in N-and O-linked glycosylation. These results indicate that SAC1 organizes PI(4)P distribution between the Golgi complex and the TGN, which is instrumental for resident enzyme partitioning and Golgi morphology.
Figures

Similar articles
-
The activity of Sac1 across ER-TGN contact sites requires the four-phosphate-adaptor-protein-1.J Cell Biol. 2019 Mar 4;218(3):783-797. doi: 10.1083/jcb.201812021. Epub 2019 Jan 18. J Cell Biol. 2019. PMID: 30659099 Free PMC article.
-
Integration of Golgi trafficking and growth factor signaling by the lipid phosphatase SAC1.J Cell Biol. 2008 Feb 25;180(4):803-12. doi: 10.1083/jcb.200708109. J Cell Biol. 2008. PMID: 18299350 Free PMC article.
-
Accumulation of PtdIns(4)P at the Golgi mediated by reversible oxidation of the PtdIns(4)P phosphatase Sac1 by H2O2.Free Radic Biol Med. 2019 Jan;130:426-435. doi: 10.1016/j.freeradbiomed.2018.11.008. Epub 2018 Nov 16. Free Radic Biol Med. 2019. PMID: 30448513
-
Growth and metabolic control of lipid signalling at the Golgi.Biochem Soc Trans. 2012 Feb;40(1):205-9. doi: 10.1042/BST20110637. Biochem Soc Trans. 2012. PMID: 22260691 Review.
-
Protein localization in the plant Golgi apparatus and the trans-Golgi network.Cell Mol Life Sci. 2004 Jan;61(2):159-71. doi: 10.1007/s00018-003-3354-7. Cell Mol Life Sci. 2004. PMID: 14745495 Free PMC article. Review.
Cited by
-
Golgi membrane dynamics and lipid metabolism.Curr Biol. 2012 May 22;22(10):R414-24. doi: 10.1016/j.cub.2012.03.004. Curr Biol. 2012. PMID: 22625862 Free PMC article. Review.
-
Regulation of V-ATPase Activity and Organelle pH by Phosphatidylinositol Phosphate Lipids.Front Cell Dev Biol. 2020 Jun 23;8:510. doi: 10.3389/fcell.2020.00510. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32656214 Free PMC article. Review.
-
Tied up: Does altering phosphoinositide-mediated membrane trafficking influence neurodegenerative disease phenotypes?J Genet. 2018 Jul;97(3):753-771. J Genet. 2018. PMID: 30027907 Review.
-
The activity of Sac1 across ER-TGN contact sites requires the four-phosphate-adaptor-protein-1.J Cell Biol. 2019 Mar 4;218(3):783-797. doi: 10.1083/jcb.201812021. Epub 2019 Jan 18. J Cell Biol. 2019. PMID: 30659099 Free PMC article.
-
The first transmembrane domain of lipid phosphatase SAC1 promotes Golgi localization.PLoS One. 2013 Aug 1;8(8):e71112. doi: 10.1371/journal.pone.0071112. Print 2013. PLoS One. 2013. PMID: 23936490 Free PMC article.
References
-
- Wang YJ, Wang J, Sun HQ, Martinez M, Sun YX, Macia E, Kirchhausen T, Albanesi JP, Roth MG, Yin HL. Phosphatidylinositol 4 phosphate regulates targeting of clathrin adaptor AP-1 complexes to the Golgi. Cell. 2003;114:299–310. - PubMed
-
- Hanada K, Kumagai K, Yasuda S, Miura Y, Kawano M, Fukasawa M, Nishijima M. Molecular machinery for non-vesicular trafficking of ceramide. Nature. 2003;426:803–809. - PubMed
-
- D’Angelo G, Polishchuk E, Di Tullio G, Santoro M, Di Campli A, Godi A, West G, Bielawski J, Chuang CC, van der Spoel AC, Platt FM, Hannun YA, Polishchuk R, Mattjus P, De Matteis MA. Glycosphingolipid synthesis requires FAPP2 transfer of glucosylceramide. Nature. 2007;449:62–67. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

