NIP/DuoxA is essential for Drosophila embryonic development and regulates oxidative stress response
- PMID: 20567495
- PMCID: PMC2878171
- DOI: 10.7150/ijbs.6.252
NIP/DuoxA is essential for Drosophila embryonic development and regulates oxidative stress response
Abstract
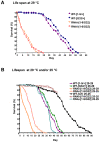
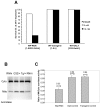
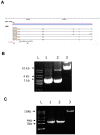
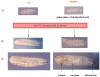
NIP/DuoxA, originally cloned as a protein capable of binding to the cell fate determinant Numb in Drosophila, was recently identified as a modulator of reactive oxygen species (ROS) production in mammalian systems. Despite biochemical and cellular studies that link NIP/DuoxA to the generation of ROS through the dual oxidase (Duox) enzyme, the in vivo function of NIP/DuoxA has not been characterized to date. Here we report a genetic and functional characterization of nip in Drosophila melanogaster. We show that nip is essential for Drosophila development as nip null mutants die at the 1(st) larval instar. Expression of UAS-nip, but not UAS-Duox, rescued the lethality. To understand the function of nip beyond the early larval stage, we generated GAL4 inducible UAS-RNAi transgenes. da(G32)-GAL4 driven, ubiquitous RNAi-mediated silencing of nip led to profound abnormality in pre-adult development, crinkled wing and markedly reduced lifespan at 29 degrees C. Compared to wild type flies, da-GAL4 induced nip-RNAi transgenic flies exhibited significantly reduced ability to survive under oxidative stress and displayed impaired mitochondrial aconitase function. Our work provides in vivo evidence for a critical role for nip in the development and oxidative stress response in Drosophila.
Keywords: Numb Interacting protein; dual oxidase maturation factor; embryonic development; oxidative stress..
Conflict of interest statement
Conflict of Interest: The authors have declared that no conflict of interest exists.
Figures

Similar articles
-
Essential role of Duox in stabilization of Drosophila wing.J Biol Chem. 2011 Sep 23;286(38):33244-51. doi: 10.1074/jbc.M111.263178. Epub 2011 Jul 30. J Biol Chem. 2011. PMID: 21808060 Free PMC article.
-
Dual Oxidase, a Hydrogen-Peroxide-Producing Enzyme, Regulates Neuronal Oxidative Damage and Animal Lifespan in Drosophila melanogaster.Cells. 2022 Jun 29;11(13):2059. doi: 10.3390/cells11132059. Cells. 2022. PMID: 35805145 Free PMC article.
-
Over-expression of human clusterin increases stress resistance and extends lifespan in Drosophila melanogaster.Biochem Biophys Res Commun. 2012 Apr 20;420(4):851-6. doi: 10.1016/j.bbrc.2012.03.087. Epub 2012 Mar 24. Biochem Biophys Res Commun. 2012. PMID: 22465014
-
Function of Lipid Storage Droplet 1 (Lsd1) in Wing Development of Drosophila melanogaster.Int J Mol Sci. 2016 Apr 29;17(5):648. doi: 10.3390/ijms17050648. Int J Mol Sci. 2016. PMID: 27136547 Free PMC article.
-
Role of oxidative stress in Drosophila aging.Mutat Res. 1992 Sep;275(3-6):267-79. doi: 10.1016/0921-8734(92)90031-j. Mutat Res. 1992. PMID: 1383769 Review.
Cited by
-
The Drosophila Duox maturation factor is a key component of a positive feedback loop that sustains regeneration signaling.PLoS Genet. 2017 Jul 28;13(7):e1006937. doi: 10.1371/journal.pgen.1006937. eCollection 2017 Jul. PLoS Genet. 2017. PMID: 28753614 Free PMC article.
-
Paradoxical roles of dual oxidases in cancer biology.Free Radic Biol Med. 2017 Sep;110:117-132. doi: 10.1016/j.freeradbiomed.2017.05.024. Epub 2017 May 31. Free Radic Biol Med. 2017. PMID: 28578013 Free PMC article. Review.
-
Calcium flashes orchestrate the wound inflammatory response through DUOX activation and hydrogen peroxide release.Curr Biol. 2013 Mar 4;23(5):424-9. doi: 10.1016/j.cub.2013.01.058. Epub 2013 Feb 7. Curr Biol. 2013. PMID: 23394834 Free PMC article.
-
Essential role of Duox in stabilization of Drosophila wing.J Biol Chem. 2011 Sep 23;286(38):33244-51. doi: 10.1074/jbc.M111.263178. Epub 2011 Jul 30. J Biol Chem. 2011. PMID: 21808060 Free PMC article.
-
Alternative oxidase confers nutritional limitation on Drosophila development.J Exp Zool A Ecol Integr Physiol. 2019 Jul;331(6):341-356. doi: 10.1002/jez.2274. Epub 2019 Jun 20. J Exp Zool A Ecol Integr Physiol. 2019. PMID: 31218852 Free PMC article.
References
-
- Uemura T, Shepherd S, Ackerman L, Jan LY, Jan YN. numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell. 1989;58(2):349–360. - PubMed
-
- Knoblich JA, Jan LY, Jan YN. Asymmetric segregation of Numb and Prospero during cell division. Nature. 1995;377(6550):624–627. - PubMed
-
- Knoblich JA. Asymmetric cell division during animal development. Nat Rev Mol Cell Biol. 2001;2(1):11–20. - PubMed
-
- Li SC, Zwahlen C, Vincent SJ, McGlade CJ, Kay LE, Pawson T, Forman-Kay JD. Structure of a Numb PTB domain-peptide complex suggests a basis for diverse binding specificity. Nat Struct Biol. 1998;5(12):1075–1083. - PubMed
-
- Schlessinger J, Lemmon MA. SH2 and PTB domains in tyrosine kinase signaling. Sci STKE. 2003;2003(191):RE12. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

