GTP-independent tRNA delivery to the ribosomal P-site by a novel eukaryotic translation factor
- PMID: 20566627
- PMCID: PMC2930676
- DOI: 10.1074/jbc.M110.119693
GTP-independent tRNA delivery to the ribosomal P-site by a novel eukaryotic translation factor
Abstract
During translation, aminoacyl-tRNAs are delivered to the ribosome by specialized GTPases called translation factors. Here, we report the tRNA binding to the P-site of 40 S ribosomes by a novel GTP-independent factor eIF2D isolated from mammalian cells. The binding of tRNA(i)(Met) occurs after the AUG codon finds its position in the P-site of 40 S ribosomes, the situation that takes place during initiation complex formation on the hepatitis C virus internal ribosome entry site or on some other specific RNAs (leaderless mRNA and A-rich mRNAs with relaxed scanning dependence). Its activity in tRNA binding with 40 S subunits does not require the presence of the aminoacyl moiety. Moreover, the factor possesses the unique ability to deliver non-Met (elongator) tRNAs into the P-site of the 40 S subunit. The corresponding gene is found in all eukaryotes and includes an SUI1 domain present also in translation initiation factor eIF1. The versatility of translation initiation strategies in eukaryotes is discussed.
Figures

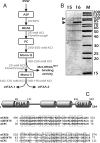
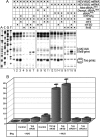
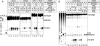
Similar articles
-
β-Hairpin loop of eukaryotic initiation factor 1 (eIF1) mediates 40 S ribosome binding to regulate initiator tRNA(Met) recruitment and accuracy of AUG selection in vivo.J Biol Chem. 2013 Sep 20;288(38):27546-27562. doi: 10.1074/jbc.M113.498642. Epub 2013 Jul 26. J Biol Chem. 2013. PMID: 23893413 Free PMC article.
-
Yeast initiator tRNA identity elements cooperate to influence multiple steps of translation initiation.RNA. 2006 May;12(5):751-64. doi: 10.1261/rna.2263906. Epub 2006 Mar 24. RNA. 2006. PMID: 16565414 Free PMC article.
-
Hepatitis C Virus Translation Regulation.Int J Mol Sci. 2020 Mar 27;21(7):2328. doi: 10.3390/ijms21072328. Int J Mol Sci. 2020. PMID: 32230899 Free PMC article. Review.
-
Activities of Ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling.Genes Dev. 2010 Aug 15;24(16):1787-801. doi: 10.1101/gad.1957510. Genes Dev. 2010. PMID: 20713520 Free PMC article.
-
Molecular mechanism of scanning and start codon selection in eukaryotes.Microbiol Mol Biol Rev. 2011 Sep;75(3):434-67, first page of table of contents. doi: 10.1128/MMBR.00008-11. Microbiol Mol Biol Rev. 2011. PMID: 21885680 Free PMC article. Review.
Cited by
-
Tma64/eIF2D, Tma20/MCT-1, and Tma22/DENR Recycle Post-termination 40S Subunits In Vivo.Mol Cell. 2018 Sep 6;71(5):761-774.e5. doi: 10.1016/j.molcel.2018.07.028. Epub 2018 Aug 23. Mol Cell. 2018. PMID: 30146315 Free PMC article.
-
A C. elegans model of C9orf72-associated ALS/FTD uncovers a conserved role for eIF2D in RAN translation.Nat Commun. 2021 Oct 15;12(1):6025. doi: 10.1038/s41467-021-26303-x. Nat Commun. 2021. PMID: 34654821 Free PMC article.
-
IRES-mediated translation of cellular messenger RNA operates in eIF2α- independent manner during stress.Nucleic Acids Res. 2012 Jan;40(2):541-52. doi: 10.1093/nar/gkr701. Epub 2011 Sep 14. Nucleic Acids Res. 2012. PMID: 21917851 Free PMC article.
-
The RNA fold interactome of evolutionary conserved RNA structures in S. cerevisiae.Nat Commun. 2020 Jun 3;11(1):2789. doi: 10.1038/s41467-020-16555-4. Nat Commun. 2020. PMID: 32493961 Free PMC article.
-
The eIF2A knockout mouse.Cell Cycle. 2016 Nov 16;15(22):3115-3120. doi: 10.1080/15384101.2016.1237324. Epub 2016 Sep 29. Cell Cycle. 2016. PMID: 27686860 Free PMC article.
References
-
- Dever T. E., Roll-Mecak A., Choi S. K., Lee J. H., Cao C., Shin B. S., Burley S. K. (2001) Cold Spring Harbor Symp. Quant. Biol. 66, 417–424 - PubMed
-
- Pestova T. V., Lomakin I. B., Lee J. H., Choi S. K., Dever T. E., Hellen C. U. (2000) Nature 403, 332–335 - PubMed
-
- Ron D., Harding H. P. (2006) in Translational Control in Biology and Medicine (Mathews M. B., Sonenberg N., Hershey J. W., eds) pp. 345–368, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
-
- Terenin I. M., Dmitriev S. E., Andreev D. E., Shatsky I. N. (2008) Nat. Struct. Mol. Biol. 15, 836–841 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

