A switch from a gradient to a threshold mode in the regulation of a transcriptional cascade promotes robust execution of meiosis in budding yeast
- PMID: 20543984
- PMCID: PMC2882377
- DOI: 10.1371/journal.pone.0011005
A switch from a gradient to a threshold mode in the regulation of a transcriptional cascade promotes robust execution of meiosis in budding yeast
Abstract
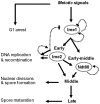
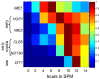
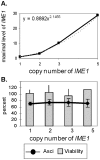
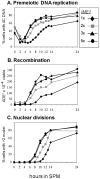
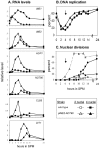
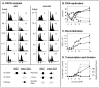
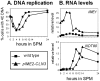
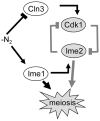
Tight regulation of developmental pathways is of critical importance to all organisms, and is achieved by a transcriptional cascade ensuring the coordinated expression of sets of genes. We aimed to explore whether a strong signal is required to enter and complete a developmental pathway, by using meiosis in budding yeast as a model. We demonstrate that meiosis in budding yeast is insensitive to drastic changes in the levels of its consecutive positive regulators (Ime1, Ime2, and Ndt80). Entry into DNA replication is not correlated with the time of transcription of the early genes that regulate this event. Entry into nuclear division is directly regulated by the time of transcription of the middle genes, as premature transcription of their activator NDT80, leads to a premature entry into the first meiotic division, and loss of coordination between DNA replication and nuclear division. We demonstrate that Cdk1/Cln3 functions as a negative regulator of Ime2, and that ectopic expression of Cln3 delays entry into nuclear division as well as NDT80 transcription. Because Ime2 functions as a positive regulator for premeiotic DNA replication and NDT80 transcription, as well as a negative regulator of Cdk/Cln, we suggest that a double negative feedback loop between Ime2 and Cdk1/Cln3 promotes a bistable switch from the cell cycle to meiosis. Moreover, our results suggest a regulatory mode switch that ensures robust meiosis as the transcription of the early meiosis-specific genes responds in a graded mode to Ime1 levels, whereas that of the middle and late genes as well as initiation of DNA replication, are regulated in a threshold mode.
Conflict of interest statement
Figures

Similar articles
-
Transcriptional regulation of meiosis in budding yeast.Int Rev Cytol. 2003;224:111-71. doi: 10.1016/s0074-7696(05)24004-4. Int Rev Cytol. 2003. PMID: 12722950 Review.
-
The Cdk1 and Ime2 protein kinases trigger exit from meiotic prophase in Saccharomyces cerevisiae by inhibiting the Sum1 transcriptional repressor.Mol Cell Biol. 2010 Jun;30(12):2996-3003. doi: 10.1128/MCB.01682-09. Epub 2010 Apr 12. Mol Cell Biol. 2010. PMID: 20385771 Free PMC article.
-
Positive and negative feedback loops affect the transcription of IME1, a positive regulator of meiosis in Saccharomyces cerevisiae.Dev Genet. 1995;16(3):219-28. doi: 10.1002/dvg.1020160302. Dev Genet. 1995. PMID: 7796531
-
GCN5-dependent histone H3 acetylation and RPD3-dependent histone H4 deacetylation have distinct, opposing effects on IME2 transcription, during meiosis and during vegetative growth, in budding yeast.Proc Natl Acad Sci U S A. 1999 Jun 8;96(12):6835-40. doi: 10.1073/pnas.96.12.6835. Proc Natl Acad Sci U S A. 1999. PMID: 10359799 Free PMC article.
-
The Sum1/Ndt80 transcriptional switch and commitment to meiosis in Saccharomyces cerevisiae.Microbiol Mol Biol Rev. 2012 Mar;76(1):1-15. doi: 10.1128/MMBR.05010-11. Microbiol Mol Biol Rev. 2012. PMID: 22390969 Free PMC article. Review.
Cited by
-
Vir1p, the yeast homolog of virilizer, is required for mRNA m6A methylation and meiosis.Genetics. 2023 May 4;224(1):iyad043. doi: 10.1093/genetics/iyad043. Genetics. 2023. PMID: 36930734 Free PMC article.
-
The roles of the catalytic and noncatalytic activities of Rpd3L and Rpd3S in the regulation of gene transcription in yeast.PLoS One. 2013 Dec 17;8(12):e85088. doi: 10.1371/journal.pone.0085088. eCollection 2013. PLoS One. 2013. PMID: 24358376 Free PMC article.
-
Nutritional control of growth and development in yeast.Genetics. 2012 Sep;192(1):73-105. doi: 10.1534/genetics.111.135731. Genetics. 2012. PMID: 22964838 Free PMC article. Review.
-
SPO24 is a transcriptionally dynamic, small ORF-encoding locus required for efficient sporulation in Saccharomyces cerevisiae.PLoS One. 2014 Aug 15;9(8):e105058. doi: 10.1371/journal.pone.0105058. eCollection 2014. PLoS One. 2014. PMID: 25127041 Free PMC article.
-
Multiple MAPK cascades regulate the transcription of IME1, the master transcriptional activator of meiosis in Saccharomyces cerevisiae.PLoS One. 2013 Nov 13;8(11):e78920. doi: 10.1371/journal.pone.0078920. eCollection 2013. PLoS One. 2013. PMID: 24236068 Free PMC article.
References
-
- Freeman M. Feedback control of intercellular signalling in development. Nature. 2000;408:313–319. - PubMed
-
- Ferrell JE., Jr Self-perpetuating states in signal transduction: positive feedback, double-negative feedback and bistability. Curr Opin Cell Biol. 2002;14:140–148. - PubMed
-
- Baugh LR, Hill AA, Claggett JM, Hill-Harfe K, Wen JC, et al. The homeodomain protein PAL-1 specifies a lineage-specific regulatory network in the C. elegans embryo. Development. 2005;132:1843–1854. - PubMed
-
- Marine JC, Topham DJ, McKay C, Wang D, Parganas E, et al. SOCS1 deficiency causes a lymphocyte-dependent perinatal lethality. Cell. 1999;98:609–616. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

