Expanding knowledge of P3 proteins in the poliovirus lifecycle
- PMID: 20521933
- PMCID: PMC2904470
- DOI: 10.2217/fmb.10.40
Expanding knowledge of P3 proteins in the poliovirus lifecycle
Abstract
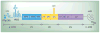
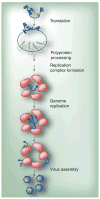
Poliovirus is the most extensively studied member of the order Picornavirales, which contains numerous medical, veterinary and agricultural pathogens. The picornavirus genome encodes a single polyprotein that is divided into three regions: P1, P2 and P3. P3 proteins are known to participate more directly in genome replication, for example by containing the viral RNA-dependent RNA polymerase (RdRp or 3Dpol), among several other proteins and enzymes. We will review recent data that provide new insight into the structure, function and mechanism of P3 proteins and their complexes, which are required for initiation of genome replication. Replication of poliovirus genomes occurs within macromolecular complexes, containing viral RNA, viral proteins and host-cell membranes, collectively referred to as replication complexes. P2 proteins clearly contribute to interactions with the host cell that are required for virus multiplication, including formation of replication complexes. We will discuss recent data that suggest a role for P3 proteins in formation of replication complexes. Among the least understood steps of the poliovirus lifecycle is encapsidation of genomic RNA. We will also describe data that suggest a role for P3 proteins in this step.
Figures


Similar articles
-
Inherent instability of poliovirus genomes containing two internal ribosome entry site (IRES) elements supports a role for the IRES in encapsidation.J Virol. 2000 Sep;74(18):8335-42. doi: 10.1128/jvi.74.18.8335-8342.2000. J Virol. 2000. PMID: 10954532 Free PMC article.
-
An efficient trans complementation system for in vivo replication of defective poliovirus mutants.J Virol. 2024 Jul 23;98(7):e0052324. doi: 10.1128/jvi.00523-24. Epub 2024 Jun 5. J Virol. 2024. PMID: 38837378 Free PMC article.
-
Cellular protein modification by poliovirus: the two faces of poly(rC)-binding protein.J Virol. 2007 Sep;81(17):8919-32. doi: 10.1128/JVI.01013-07. Epub 2007 Jun 20. J Virol. 2007. PMID: 17581994 Free PMC article.
-
IRES-controlled protein synthesis and genome replication of poliovirus.Arch Virol Suppl. 1994;9:279-89. doi: 10.1007/978-3-7091-9326-6_28. Arch Virol Suppl. 1994. PMID: 8032259 Review.
-
Involvement of cellular membrane traffic proteins in poliovirus replication.Cell Cycle. 2007 Jan 1;6(1):36-8. doi: 10.4161/cc.6.1.3683. Epub 2007 Jan 11. Cell Cycle. 2007. PMID: 17245115 Review.
Cited by
-
RNA synthetic mechanisms employed by diverse families of RNA viruses.Wiley Interdiscip Rev RNA. 2013 Jul-Aug;4(4):351-67. doi: 10.1002/wrna.1164. Epub 2013 Apr 18. Wiley Interdiscip Rev RNA. 2013. PMID: 23606593 Free PMC article. Review.
-
Initiation of protein-primed picornavirus RNA synthesis.Virus Res. 2015 Aug 3;206:12-26. doi: 10.1016/j.virusres.2014.12.028. Epub 2015 Jan 12. Virus Res. 2015. PMID: 25592245 Free PMC article. Review.
-
The genome-linked protein VPg of vertebrate viruses - a multifaceted protein.Curr Opin Virol. 2011 Nov;1(5):355-62. doi: 10.1016/j.coviro.2011.09.003. Epub 2011 Oct 7. Curr Opin Virol. 2011. PMID: 22440837 Free PMC article. Review.
-
Current Challenges for the Effective Management of the COVID-19 Pandemic.Adv Exp Med Biol. 2021;1353:131-149. doi: 10.1007/978-3-030-85113-2_8. Adv Exp Med Biol. 2021. PMID: 35137372
-
Viral targets of acylguanidines.Drug Discov Today. 2012 Sep;17(17-18):1039-43. doi: 10.1016/j.drudis.2012.05.002. Epub 2012 May 8. Drug Discov Today. 2012. PMID: 22580299 Free PMC article. Review.
References
-
- Racaniello VR. Picornaviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. 5. Lippincott Williams & Wilkins; Philadelphia, PA, USA: 2007. pp. 765–838.
-
- Le Gall O, Christian P, Fauquet CM, et al. Picornavirales, a proposed order of positive-sense single-stranded RNA viruses with a pseudo-T = 3 virion architecture. Arch Virol. 2008;153(4):715–727. - PubMed
-
- Paul AV, Belov GA, Ehrenfeld E, Wimmer E. Model of picornavirus RNA replication. In: Cameron CE, Götte M, Raney KD, editors. Viral Genome Replication. 1. Vol. 3. Springer; Philadelphia, PA: 2009. p. 23.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
