Wss1 is a SUMO-dependent isopeptidase that interacts genetically with the Slx5-Slx8 SUMO-targeted ubiquitin ligase
- PMID: 20516210
- PMCID: PMC2916399
- DOI: 10.1128/MCB.01649-09
Wss1 is a SUMO-dependent isopeptidase that interacts genetically with the Slx5-Slx8 SUMO-targeted ubiquitin ligase
Abstract
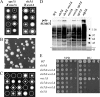
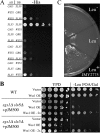
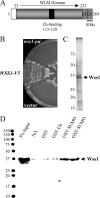
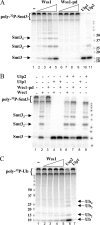
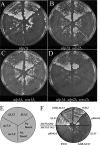
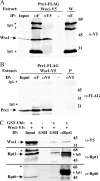
Protein sumoylation plays an important but poorly understood role in controlling genome integrity. In Saccharomyces cerevisiae, the Slx5-Slx8 SUMO-targeted Ub ligase appears to be needed to ubiquitinate sumoylated proteins that arise in the absence of the Sgs1 DNA helicase. WSS1, a high-copy-number suppressor of a mutant SUMO, was implicated in this pathway because it shares phenotypes with SLX5-SLX8 mutants, including a wss1Delta sgs1Delta synthetic-fitness defect. Here we show that Wss1, a putative metalloprotease, physically binds SUMO and displays in vitro isopeptidase activity on poly-SUMO chains. Like that of SLX5, overexpression of WSS1 suppresses sgs1Delta slx5Delta lethality and the ulp1ts growth defect. Interestingly, although Wss1 is relatively inactive on ubiquitinated substrates and poly-Ub chains, it efficiently deubiquitinates a Ub-SUMO isopeptide conjugate and a Ub-SUMO fusion protein. Wss1 was further implicated in Ub metabolism on the basis of its physical association with proteasomal subunits. The results suggest that Wss1 is a SUMO-dependent isopeptidase that acts on sumoylated substrates as they undergo proteasomal degradation.
Figures



Similar articles
-
Genetic evidence that polysumoylation bypasses the need for a SUMO-targeted Ub ligase.Genetics. 2011 Jan;187(1):73-87. doi: 10.1534/genetics.110.124347. Epub 2010 Nov 8. Genetics. 2011. PMID: 21059884 Free PMC article.
-
Activation of the Slx5-Slx8 ubiquitin ligase by poly-small ubiquitin-like modifier conjugates.J Biol Chem. 2008 Jul 18;283(29):19912-21. doi: 10.1074/jbc.M802690200. Epub 2008 May 22. J Biol Chem. 2008. PMID: 18499666 Free PMC article.
-
A Lysine Desert Protects a Novel Domain in the Slx5-Slx8 SUMO Targeted Ub Ligase To Maintain Sumoylation Levels in Saccharomyces cerevisiae.Genetics. 2017 Aug;206(4):1807-1821. doi: 10.1534/genetics.117.202697. Epub 2017 May 26. Genetics. 2017. PMID: 28550017 Free PMC article.
-
SUMO-targeted ubiquitin ligases.Biochim Biophys Acta. 2014 Jan;1843(1):75-85. doi: 10.1016/j.bbamcr.2013.08.022. Epub 2013 Sep 7. Biochim Biophys Acta. 2014. PMID: 24018209 Review.
-
Genome maintenance in Saccharomyces cerevisiae: the role of SUMO and SUMO-targeted ubiquitin ligases.Nucleic Acids Res. 2017 Mar 17;45(5):2242-2261. doi: 10.1093/nar/gkw1369. Nucleic Acids Res. 2017. PMID: 28115630 Free PMC article. Review.
Cited by
-
SUMO: a multifaceted modifier of chromatin structure and function.Dev Cell. 2013 Jan 14;24(1):1-12. doi: 10.1016/j.devcel.2012.11.020. Dev Cell. 2013. PMID: 23328396 Free PMC article. Review.
-
The Nup84 complex coordinates the DNA damage response to warrant genome integrity.Nucleic Acids Res. 2019 May 7;47(8):4054-4067. doi: 10.1093/nar/gkz066. Nucleic Acids Res. 2019. PMID: 30715474 Free PMC article.
-
Update on sumoylation: defining core components of the plant SUMO conjugation system by phylogenetic comparison.New Phytol. 2012 Jul;195(1):23-31. doi: 10.1111/j.1469-8137.2012.04135.x. New Phytol. 2012. PMID: 22799003 Free PMC article. Review.
-
The yeast homologue of the microtubule-associated protein Lis1 interacts with the sumoylation machinery and a SUMO-targeted ubiquitin ligase.Mol Biol Cell. 2012 Dec;23(23):4552-66. doi: 10.1091/mbc.E12-03-0195. Epub 2012 Oct 3. Mol Biol Cell. 2012. PMID: 23034179 Free PMC article.
-
Targeting DNA-Protein Crosslinks via Post-Translational Modifications.Front Mol Biosci. 2022 Jul 4;9:944775. doi: 10.3389/fmolb.2022.944775. eCollection 2022. Front Mol Biosci. 2022. PMID: 35860355 Free PMC article. Review.
References
-
- Bachant, J., A. Alcasabas, Y. Blat, N. Kleckner, and S. J. Elledge. 2002. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol. Cell 9:1169-1182. - PubMed
-
- Branzei, D., J. Sollier, G. Liberi, X. Zhao, D. Maeda, M. Seki, T. Enomoto, K. Ohta, and M. Foiani. 2006. Ubc9- and Mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell 127:509-522. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
