Quality control of inclusion bodies in Escherichia coli
- PMID: 20509924
- PMCID: PMC2893105
- DOI: 10.1186/1475-2859-9-41
Quality control of inclusion bodies in Escherichia coli
Abstract
Background: Bacterial inclusion bodies (IBs) are key intermediates for protein production. Their quality affects the refolding yield and further purification. Recent functional and structural studies have revealed that IBs are not dead-end aggregates but undergo dynamic changes, including aggregation, refunctionalization of the protein and proteolysis. Both, aggregation of the folding intermediates and turnover of IBs are influenced by the cellular situation and a number of well-studied chaperones and proteases are included. IBs mostly contain only minor impurities and are relatively homogenous.
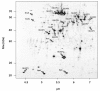
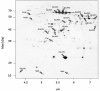
Results: IBs of alpha-glucosidase of Saccharomyces cerevisiae after overproduction in Escherichia coli contain a large amount of (at least 12 different) major product fragments, as revealed by two-dimensional polyacrylamide gel electrophoresis (2D PAGE). Matrix-Assisted-Laser-Desorption/Ionization-Time-Of-Flight Mass-Spectrometry (MALDI-ToF MS) identification showed that these fragments contain either the N- or the C-terminus of the protein, therefore indicate that these IBs are at least partially created by proteolytic action. Expression of alpha-glucosidase in single knockout mutants for the major proteases ClpP, Lon, OmpT and FtsH which are known to be involved in the heat shock like response to production of recombinant proteins or to the degradation of IB proteins, clpP, lon, ompT, and ftsH did not influence the fragment pattern or the composition of the IBs. The quality of the IBs was also not influenced by the sampling time, cultivation medium (complex and mineral salt medium), production strategy (shake flask, fed-batch fermentation process), production strength (T5-lac or T7 promoter), strain background (K-12 or BL21), or addition of different protease inhibitors during IB preparation.
Conclusions: alpha-glucosidase is fragmented before aggregation, but neither by proteolytic action on the IBs by the common major proteases, nor during downstream IB preparation. Different fragments co-aggregate in the process of IB formation together with the full-length product. Other intracellular proteases than ClpP or Lon must be responsible for fragmentation. Reaggregation of protease-stable alpha-glucosidase fragments during in situ disintegration of the existing IBs does not seem to occur.
Figures


Similar articles
-
Lon and ClpP proteases participate in the physiological disintegration of bacterial inclusion bodies.J Biotechnol. 2005 Sep 23;119(2):163-71. doi: 10.1016/j.jbiotec.2005.04.006. J Biotechnol. 2005. PMID: 15967532
-
Monitoring of genes that respond to overproduction of an insoluble recombinant protein in Escherichia coli glucose-limited fed-batch fermentations.Biotechnol Bioeng. 2000 Oct 20;70(2):217-24. doi: 10.1002/1097-0290(20001020)70:2<217::aid-bit11>3.0.co;2-w. Biotechnol Bioeng. 2000. PMID: 10972933
-
Synergistic roles of HslVU and other ATP-dependent proteases in controlling in vivo turnover of sigma32 and abnormal proteins in Escherichia coli.J Bacteriol. 1997 Dec;179(23):7219-25. doi: 10.1128/jb.179.23.7219-7225.1997. J Bacteriol. 1997. PMID: 9393683 Free PMC article.
-
Comparative genomics and functional roles of the ATP-dependent proteases Lon and Clp during cytosolic protein degradation.Res Microbiol. 2004 Nov;155(9):710-9. doi: 10.1016/j.resmic.2004.06.003. Res Microbiol. 2004. PMID: 15501647 Review.
-
Proteases in bacterial pathogenesis.Res Microbiol. 2009 Nov;160(9):704-10. doi: 10.1016/j.resmic.2009.08.017. Epub 2009 Sep 22. Res Microbiol. 2009. PMID: 19778606 Review.
Cited by
-
A novel hyaluronidase from brown spider (Loxosceles intermedia) venom (Dietrich's Hyaluronidase): from cloning to functional characterization.PLoS Negl Trop Dis. 2013 May 2;7(5):e2206. doi: 10.1371/journal.pntd.0002206. Print 2013. PLoS Negl Trop Dis. 2013. PMID: 23658852 Free PMC article.
-
Structural Stabilization of Clinically Oriented Oligomeric Proteins During their Transit through Synthetic Secretory Amyloids.Adv Sci (Weinh). 2024 Jun;11(21):e2309427. doi: 10.1002/advs.202309427. Epub 2024 Mar 19. Adv Sci (Weinh). 2024. PMID: 38501900 Free PMC article.
-
Post-production protein stability: trouble beyond the cell factory.Microb Cell Fact. 2011 Aug 1;10:60. doi: 10.1186/1475-2859-10-60. Microb Cell Fact. 2011. PMID: 21806813 Free PMC article.
-
Multiple phenotypic changes associated with large-scale horizontal gene transfer.PLoS One. 2014 Jul 21;9(7):e102170. doi: 10.1371/journal.pone.0102170. eCollection 2014. PLoS One. 2014. PMID: 25048697 Free PMC article.
-
Production strategies for active heme-containing peroxidases from E. coli inclusion bodies - a review.Biotechnol Rep (Amst). 2016 Mar 24;10:75-83. doi: 10.1016/j.btre.2016.03.005. eCollection 2016 Jun. Biotechnol Rep (Amst). 2016. PMID: 28352527 Free PMC article. Review.
References
-
- Mayer M, Buchner J. Refolding of inclusion body proteins. Methods Mol Med. 2004;94:239–254. - PubMed
-
- Rudolph R. In: Principles and Practice of Protein Folding. Cleland JL, Craik CS, editor. New York: J. Wiley & Sons Inc.; 1996. Successful protein folding on an industrial scale.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

