Decoded calreticulin-deficient embryonic stem cell transcriptome resolves latent cardiophenotype
- PMID: 20506533
- PMCID: PMC3129684
- DOI: 10.1002/stem.447
Decoded calreticulin-deficient embryonic stem cell transcriptome resolves latent cardiophenotype
Abstract
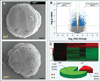
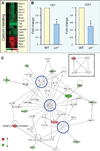
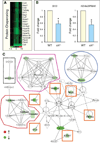
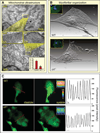
Genomic perturbations that challenge normal signaling at the pluripotent stage may trigger unforeseen ontogenic aberrancies. Anticipatory systems biology identification of transcriptome landscapes that underlie latent phenotypes would offer molecular diagnosis before the onset of symptoms. The purpose of this study was to assess the impact of calreticulin-deficient embryonic stem cell transcriptomes on molecular functions and physiological systems. Bioinformatic surveillance of calreticulin-null stem cells, a monogenic insult model, diagnosed a disruption in transcriptome dynamics, which re-prioritized essential cellular functions. Calreticulin-calibrated signaling axes were uncovered, and network-wide cartography of undifferentiated stem cell transcripts suggested cardiac manifestations. Calreticulin-deficient stem cell-derived cardiac cells verified disorganized sarcomerogenesis, mitochondrial paucity, and cytoarchitectural aberrations to validate calreticulin-dependent network forecasts. Furthermore, magnetic resonance imaging and histopathology detected a ventricular septal defect, revealing organogenic manifestation of calreticulin deletion. Thus, bioinformatic deciphering of a primordial calreticulin-deficient transcriptome decoded at the pluripotent stem cell stage a reconfigured multifunctional molecular registry to anticipate predifferentiation susceptibility toward abnormal cardiophenotype.
Conflict of interest statement
The authors indicate no potential conflicts of interest.
Figures


Similar articles
-
Systems biology surveillance decrypts pathological transcriptome remodeling.BMC Syst Biol. 2015 Jul 17;9:36. doi: 10.1186/s12918-015-0177-8. BMC Syst Biol. 2015. PMID: 26179794 Free PMC article.
-
Embryonic stem cell-derived cardiomyogenesis: a novel role for calreticulin as a regulator.Stem Cells. 2009 Jul;27(7):1507-15. doi: 10.1002/stem.85. Stem Cells. 2009. PMID: 19544459
-
Tamoxifen-inducible Cre-mediated calreticulin excision to study mouse embryonic stem cell differentiation.Stem Cells Dev. 2009 Jan-Feb;18(1):187-93. doi: 10.1089/scd.2008.0064. Stem Cells Dev. 2009. PMID: 18643752
-
Calreticulin in the heart.Mol Cell Biochem. 2004 Aug;263(1-2):137-42. Mol Cell Biochem. 2004. PMID: 15524174 Review.
-
Calreticulin is an upstream regulator of calcineurin.Biochem Biophys Res Commun. 2003 Nov 28;311(4):1173-9. doi: 10.1016/j.bbrc.2003.08.040. Biochem Biophys Res Commun. 2003. PMID: 14623303 Review.
Cited by
-
Non-Endoplasmic Reticulum-Based Calr (Calreticulin) Can Coordinate Heterocellular Calcium Signaling and Vascular Function.Arterioscler Thromb Vasc Biol. 2018 Jan;38(1):120-130. doi: 10.1161/ATVBAHA.117.309886. Epub 2017 Nov 9. Arterioscler Thromb Vasc Biol. 2018. PMID: 29122814 Free PMC article.
-
Disrupted WNT signaling in mouse embryonic stem cells in the absence of calreticulin.Stem Cell Rev Rep. 2014 Apr;10(2):191-206. doi: 10.1007/s12015-013-9488-6. Stem Cell Rev Rep. 2014. PMID: 24415131
-
Systems biology surveillance decrypts pathological transcriptome remodeling.BMC Syst Biol. 2015 Jul 17;9:36. doi: 10.1186/s12918-015-0177-8. BMC Syst Biol. 2015. PMID: 26179794 Free PMC article.
-
Systems proteomics for translational network medicine.Circ Cardiovasc Genet. 2012 Aug 1;5(4):478. doi: 10.1161/CIRCGENETICS.110.958991. Circ Cardiovasc Genet. 2012. PMID: 22896016 Free PMC article.
-
Mechanisms of protein homeostasis (proteostasis) maintain stem cell identity in mammalian pluripotent stem cells.Cell Mol Life Sci. 2018 Jan;75(2):275-290. doi: 10.1007/s00018-017-2602-1. Epub 2017 Jul 26. Cell Mol Life Sci. 2018. PMID: 28748323 Free PMC article. Review.
References
-
- Suzuki A, Raya Á, Kawakami Y, et al. Maintenance of embryonic stem cell pluripotency by Nanog-mediated reversal of mesoderm specification. Nat Clin Pract Cardiovasc Med. 2006;3:S114–S122. - PubMed
-
- Loh Y-H, Wu Q, Chew J-L, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. - PubMed
-
- Faustino RS, Terzic A. Interactome of a cardiopoietic precursor. J Cardiovasc Trans Res. 2008;1:120–126. - PubMed
-
- Komili S, Silver PA. Coupling and coordination in gene expression processes: A systems biology view. Nat Rev Genet. 2008;9:38–48. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

