Functional dissection of an intrinsically disordered protein: understanding the roles of different domains of Knr4 protein in protein-protein interactions
- PMID: 20506404
- PMCID: PMC2974829
- DOI: 10.1002/pro.418
Functional dissection of an intrinsically disordered protein: understanding the roles of different domains of Knr4 protein in protein-protein interactions
Abstract
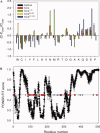
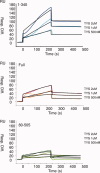
Knr4, recently characterized as an intrinsically disordered Saccharomyces cerevisiae protein, participates in cell wall formation and cell cycle regulation. It is constituted of a functional central globular core flanked by a poorly structured N-terminal and large natively unfolded C-terminal domains. Up to now, about 30 different proteins have been reported to physically interact with Knr4. Here, we used an in vivo two-hybrid system approach and an in vitro surface plasmon resonance (BIAcore) technique to compare the interaction level of different Knr4 deletion variants with given protein partners. We demonstrate the indispensability of the N-terminal domain of Knr4 for the interactions. On the other hand, presence of the unstructured C-terminal domain has a negative effect on the interaction strength. In protein interactions networks, the most highly connected proteins or "hubs" are significantly enriched in unstructured regions, and among them the transient hub proteins contain the largest and most highly flexible regions. The results presented here of our analysis of Knr4 protein suggest that these large disordered regions are not always involved in promoting the protein-protein interactions of hub proteins, but in some cases, might rather inhibit them. We propose that this type of regions could prevent unspecific protein interactions, or ensure the correct timing of occurrence of transient interactions, which may be of crucial importance for different signaling and regulation processes.
Figures


Similar articles
-
The Conserved Yeast Protein Knr4 Involved in Cell Wall Integrity Is a Multi-domain Intrinsically Disordered Protein.J Mol Biol. 2023 May 15;435(10):168048. doi: 10.1016/j.jmb.2023.168048. Epub 2023 Mar 17. J Mol Biol. 2023. PMID: 36933821
-
Structure-function analysis of Knr4/Smi1, a newly member of intrinsically disordered proteins family, indispensable in the absence of a functional PKC1-SLT2 pathway in Saccharomyces cerevisiae.Yeast. 2008 Aug;25(8):563-76. doi: 10.1002/yea.1608. Yeast. 2008. PMID: 18668512
-
Knr4 N-terminal domain controls its localization and function during sexual differentiation and vegetative growth.Yeast. 2010 Aug;27(8):563-74. doi: 10.1002/yea.1804. Yeast. 2010. PMID: 20602333
-
Knr4: a disordered hub protein at the heart of fungal cell wall signalling.Cell Microbiol. 2016 Sep;18(9):1217-27. doi: 10.1111/cmi.12618. Epub 2016 Jun 28. Cell Microbiol. 2016. PMID: 27199081 Review.
-
The 'interactome' of the Knr4/Smi1, a protein implicated in coordinating cell wall synthesis with bud emergence in Saccharomyces cerevisiae.Mol Genet Genomics. 2006 Mar;275(3):217-30. doi: 10.1007/s00438-005-0082-8. Epub 2005 Dec 16. Mol Genet Genomics. 2006. PMID: 16362369
Cited by
-
Crystallographic studies of the structured core domain of Knr4 from Saccharomyces cerevisiae.Acta Crystallogr F Struct Biol Commun. 2015 Sep;71(Pt 9):1120-4. doi: 10.1107/S2053230X15012522. Epub 2015 Aug 25. Acta Crystallogr F Struct Biol Commun. 2015. PMID: 26323295 Free PMC article.
-
Novel IM-associated protein Tim54 plays a role in the mitochondrial import of internal signal-containing proteins in Trypanosoma brucei.Biol Cell. 2021 Jan;113(1):39-57. doi: 10.1111/boc.202000054. Epub 2020 Nov 9. Biol Cell. 2021. PMID: 33084070 Free PMC article.
-
In vivo protein complex topologies: sights through a cross-linking lens.Proteomics. 2012 May;12(10):1565-75. doi: 10.1002/pmic.201100516. Proteomics. 2012. PMID: 22610688 Free PMC article. Review.
-
A novel MAP kinase-interacting protein MoSmi1 regulates development and pathogenicity in Magnaporthe oryzae.Mol Plant Pathol. 2024 Jul;25(7):e13493. doi: 10.1111/mpp.13493. Mol Plant Pathol. 2024. PMID: 39034619 Free PMC article.
-
A novel immunity system for bacterial nucleic acid degrading toxins and its recruitment in various eukaryotic and DNA viral systems.Nucleic Acids Res. 2011 Jun;39(11):4532-52. doi: 10.1093/nar/gkr036. Epub 2011 Feb 8. Nucleic Acids Res. 2011. PMID: 21306995 Free PMC article.
References
-
- Martin H, Dagkessamanskaia A, Satchanska G, Dallies N, Francois J. KNR4, a suppressor of Saccharomyces cerevisiae cwh mutants, is involved in the transcriptional control of chitin synthase genes. Microbiology. 1999;145:249–258. - PubMed
-
- Martin-Yken H, Dagkessamanskaia A, Basmaji F, Lagorce A, Francois J. The interaction of Slt2 MAP kinase with Knr4 is necessary for signalling through the cell wall integrity pathway in Saccharomyces cerevisiae. Mol Microbiol. 2003;49:23–35. - PubMed
-
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwi B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. - PubMed
-
- Dagkessamanskaia A, Martin-Yken H, Basmaji F, Briza P, Francois J. Interaction of Knr4 protein, a protein involved in cell wall synthesis, with tyrosine tRNA synthetase encoded by TYS1 in Saccharomyces cerevisiae. FEMS Microbiol Lett. 2001;200:53–58. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

